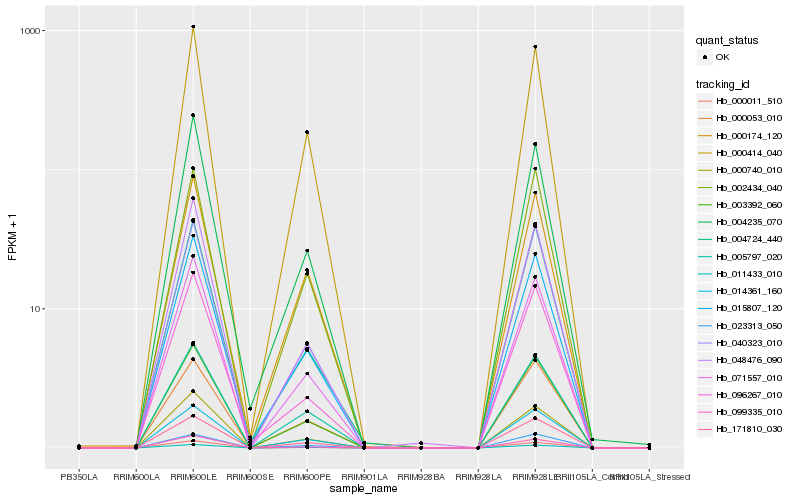
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003392_060 |
0.0 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 2 |
Hb_071557_010 |
0.0263320486 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 3 |
Hb_011433_010 |
0.0264612045 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |
| 4 |
Hb_014361_160 |
0.0277866719 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 5 |
Hb_171810_030 |
0.0300253081 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 6 |
Hb_015807_120 |
0.0328179552 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000740_010 |
0.0397580097 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_005797_020 |
0.041176437 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 9 |
Hb_040323_010 |
0.0463806101 |
- |
- |
- |
| 10 |
Hb_000414_040 |
0.046748933 |
- |
- |
hypothetical protein POPTR_0009s13250g [Populus trichocarpa] |
| 11 |
Hb_000011_510 |
0.04751582 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 12 |
Hb_004724_440 |
0.0504685079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000053_010 |
0.0520934351 |
- |
- |
hypothetical protein PRUPE_ppa017696mg [Prunus persica] |
| 14 |
Hb_099335_010 |
0.0522786391 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 15 |
Hb_002434_040 |
0.0539123239 |
- |
- |
hypothetical protein CICLE_v10022672mg [Citrus clementina] |
| 16 |
Hb_023313_050 |
0.0579399502 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein rcd1-like [Jatropha curcas] |
| 17 |
Hb_048476_090 |
0.0589760185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004235_070 |
0.0595451713 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 19 |
Hb_096267_010 |
0.0601465109 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 20 |
Hb_000174_120 |
0.0645877967 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |