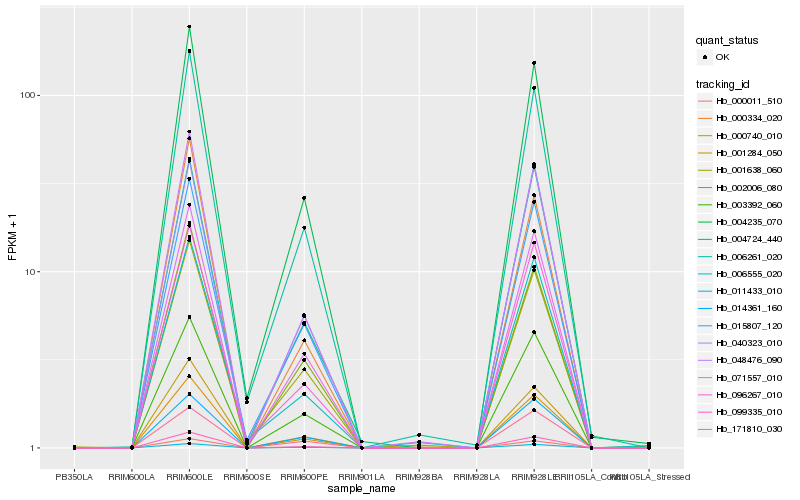
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099335_010 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 2 |
Hb_000011_510 |
0.0207967241 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 3 |
Hb_048476_090 |
0.0245508984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_071557_010 |
0.0340781577 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 5 |
Hb_000740_010 |
0.0350309814 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_096267_010 |
0.0489164516 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 7 |
Hb_003392_060 |
0.0522786391 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 8 |
Hb_004235_070 |
0.0546015099 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 9 |
Hb_006261_020 |
0.0566460005 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 10 |
Hb_015807_120 |
0.0582348455 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001284_050 |
0.0610437004 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 12 |
Hb_004724_440 |
0.0655284239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006555_020 |
0.066706477 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 14 |
Hb_171810_030 |
0.0721324902 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 15 |
Hb_040323_010 |
0.0737651086 |
- |
- |
- |
| 16 |
Hb_002006_080 |
0.074047038 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 17 |
Hb_000334_020 |
0.0767331802 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_014361_160 |
0.0778400963 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 19 |
Hb_001638_060 |
0.0779841677 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 20 |
Hb_011433_010 |
0.078048631 |
- |
- |
PREDICTED: (R)-mandelonitrile lyase-like [Populus euphratica] |