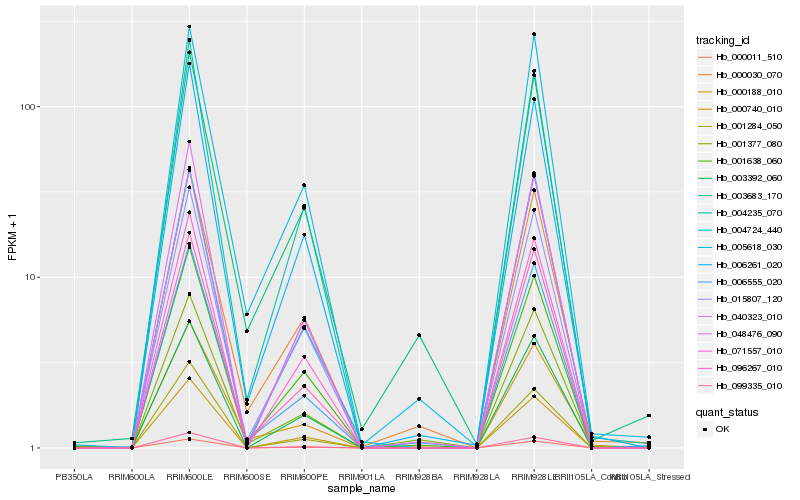
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006555_020 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 2 |
Hb_096267_010 |
0.0459433744 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 3 |
Hb_000030_070 |
0.0591845353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005618_030 |
0.0625253409 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 5 |
Hb_001377_080 |
0.0629641315 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 6 |
Hb_000011_510 |
0.0633831502 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 7 |
Hb_048476_090 |
0.0655780246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006261_020 |
0.0663843687 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 9 |
Hb_099335_010 |
0.066706477 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C2-like [Populus euphratica] |
| 10 |
Hb_001638_060 |
0.0716039573 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 11 |
Hb_003683_170 |
0.0717194995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_015807_120 |
0.0725374575 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004724_440 |
0.0729062528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004235_070 |
0.0735632469 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 15 |
Hb_071557_010 |
0.0770064221 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 16 |
Hb_000740_010 |
0.0809472308 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001284_050 |
0.0813671938 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 18 |
Hb_003392_060 |
0.0816666661 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_040323_010 |
0.0861467959 |
- |
- |
- |
| 20 |
Hb_000188_010 |
0.0864295674 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |