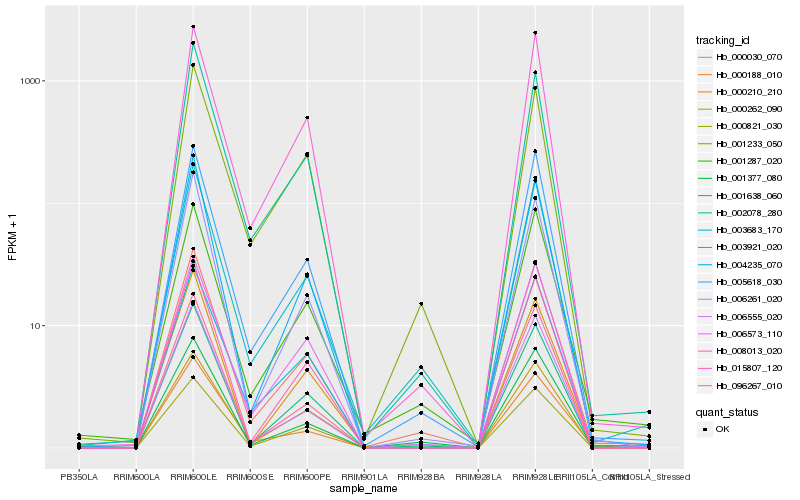
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003683_170 |
0.0359398311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005618_030 |
0.0484210455 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 4 |
Hb_001377_080 |
0.0561826447 |
- |
- |
PREDICTED: uncharacterized protein LOC105642864 [Jatropha curcas] |
| 5 |
Hb_001638_060 |
0.0565900065 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 6 |
Hb_006555_020 |
0.0591845353 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 7 |
Hb_001287_020 |
0.0620345238 |
- |
- |
SulA [Manihot esculenta] |
| 8 |
Hb_000821_030 |
0.0643755792 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-2 [Jatropha curcas] |
| 9 |
Hb_006261_020 |
0.0693582612 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 10 |
Hb_003921_020 |
0.0703675633 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 11 |
Hb_015807_120 |
0.0713372537 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008013_020 |
0.072059958 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002078_280 |
0.072674677 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 14 |
Hb_004235_070 |
0.0750523107 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 15 |
Hb_000188_010 |
0.0759557802 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 16 |
Hb_096267_010 |
0.0778734731 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 17 |
Hb_000262_090 |
0.0803694822 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 18 |
Hb_001233_050 |
0.0827880713 |
- |
- |
Magnesium-protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000210_210 |
0.0839958816 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006573_110 |
0.084901494 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |