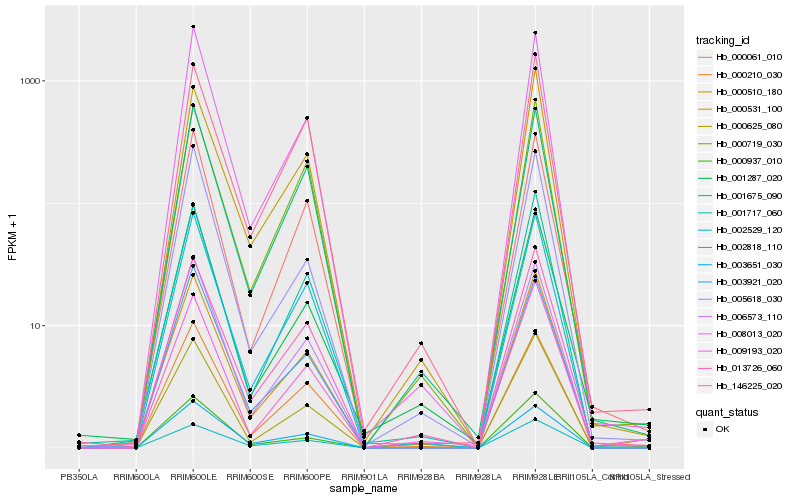
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_180 |
0.0 |
- |
- |
unknown [Lotus japonicus] |
| 2 |
Hb_000625_080 |
0.0449386838 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 3 |
Hb_013726_060 |
0.0628543484 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 4 |
Hb_002818_110 |
0.0631499895 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 5 |
Hb_008013_020 |
0.0634124995 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006573_110 |
0.0669344547 |
- |
- |
PREDICTED: uncharacterized protein PAM68-like [Jatropha curcas] |
| 7 |
Hb_009193_020 |
0.0682336685 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 8 |
Hb_001717_060 |
0.0687188581 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 9 |
Hb_000531_100 |
0.0763099454 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_003921_020 |
0.0772705253 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 11 |
Hb_000937_010 |
0.0782780412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000061_010 |
0.0787516513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000719_030 |
0.0810433541 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_005618_030 |
0.0822970603 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 15 |
Hb_002529_120 |
0.0825710672 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 16 |
Hb_000210_030 |
0.0831142178 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP6 [Jatropha curcas] |
| 17 |
Hb_003651_030 |
0.0840406931 |
- |
- |
lyase, putative [Ricinus communis] |
| 18 |
Hb_146225_020 |
0.0843470319 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 19 |
Hb_001675_090 |
0.0845346364 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001287_020 |
0.0847376454 |
- |
- |
SulA [Manihot esculenta] |