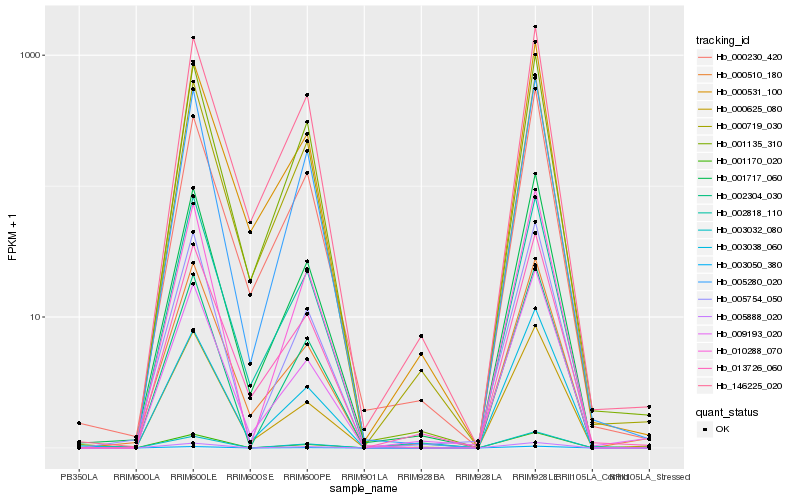
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001717_060 |
0.0 |
- |
- |
PREDICTED: xylogen-like protein 11 [Gossypium raimondii] |
| 2 |
Hb_003038_060 |
0.0503417185 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 3 |
Hb_005754_050 |
0.0550100655 |
- |
- |
PREDICTED: uncharacterized protein LOC105636679 [Jatropha curcas] |
| 4 |
Hb_000531_100 |
0.0565296639 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_005280_020 |
0.0573925604 |
- |
- |
glycine cleavage system h protein, putative [Ricinus communis] |
| 6 |
Hb_013726_060 |
0.0590857253 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 7 |
Hb_002818_110 |
0.0623677456 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 8 |
Hb_000625_080 |
0.0623813084 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001135_310 |
0.0624627478 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 10 |
Hb_000230_420 |
0.0640022613 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 11 |
Hb_010288_070 |
0.0641151103 |
transcription factor |
TF Family: MYB |
MYB-like DNA-binding domain protein [Theobroma cacao] |
| 12 |
Hb_146225_020 |
0.0664607093 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 13 |
Hb_003050_380 |
0.0677854967 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 14 |
Hb_000719_030 |
0.068310467 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000510_180 |
0.0687188581 |
- |
- |
unknown [Lotus japonicus] |
| 16 |
Hb_002304_030 |
0.0688344693 |
- |
- |
hypothetical protein JCGZ_21517 [Jatropha curcas] |
| 17 |
Hb_009193_020 |
0.0691606827 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 18 |
Hb_005888_020 |
0.0697195059 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 19 |
Hb_001170_020 |
0.0707553654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003032_080 |
0.0720499203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |