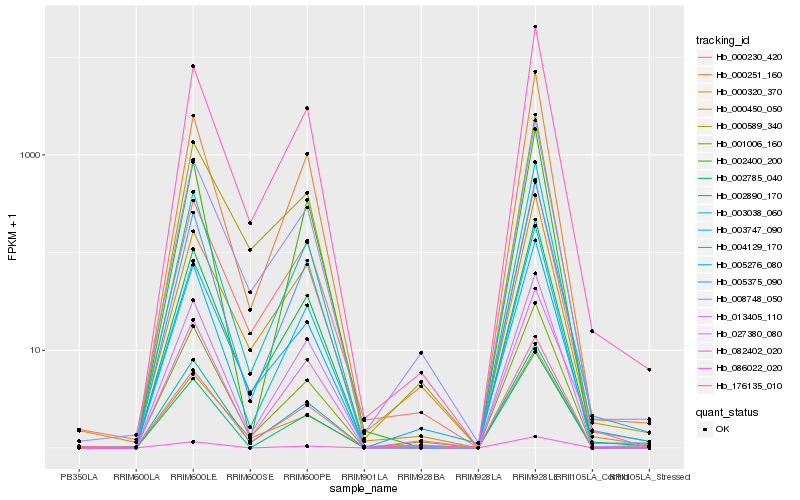
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002785_040 |
0.0 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 2 |
Hb_003747_090 |
0.0241381426 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_005375_090 |
0.0356770946 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 4 |
Hb_013405_110 |
0.0364792839 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_086022_020 |
0.0425683472 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 6 |
Hb_004129_170 |
0.0542139294 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 7 |
Hb_000320_370 |
0.0544178883 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001006_160 |
0.056680511 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_008748_050 |
0.0577369217 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_000450_050 |
0.0616696865 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 11 |
Hb_000589_340 |
0.0624228982 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 12 |
Hb_002890_170 |
0.0655835122 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 13 |
Hb_176135_010 |
0.0657480468 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_002400_200 |
0.0658042332 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 15 |
Hb_005276_080 |
0.0658685791 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 16 |
Hb_082402_020 |
0.0660261356 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 17 |
Hb_027380_080 |
0.0665621725 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 18 |
Hb_003038_060 |
0.0665911991 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 19 |
Hb_000251_160 |
0.0673543489 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 20 |
Hb_000230_420 |
0.0673870467 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |