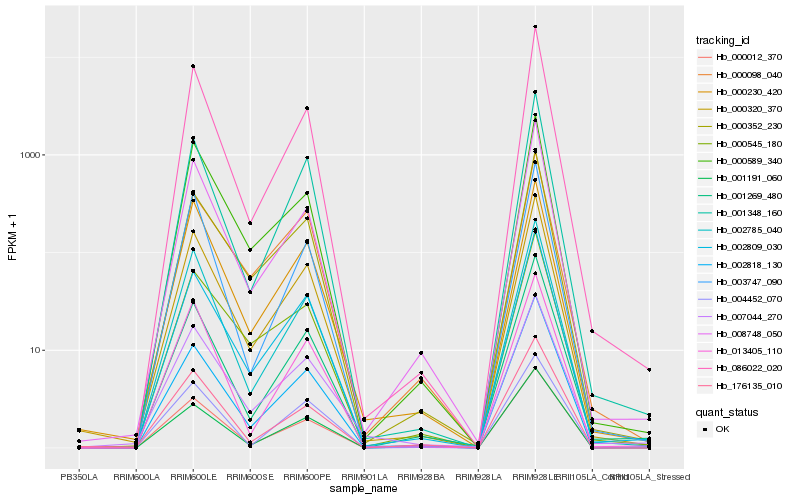
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_370 |
0.0 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002809_030 |
0.0423636811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002785_040 |
0.0544178883 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_086022_020 |
0.0556028494 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 5 |
Hb_000352_230 |
0.0571291977 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_008748_050 |
0.0600910623 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_000589_340 |
0.0646822413 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_001348_160 |
0.065126608 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000012_370 |
0.0669046976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_176135_010 |
0.067487714 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_007044_270 |
0.0686791463 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 12 |
Hb_013405_110 |
0.0691306695 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_001191_060 |
0.0708363594 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 14 |
Hb_000230_420 |
0.0711914708 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 15 |
Hb_000545_180 |
0.0730209613 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_003747_090 |
0.0735539169 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_002818_130 |
0.0737956557 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_000098_040 |
0.0743616003 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001269_480 |
0.0756339098 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004452_070 |
0.0761305964 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |