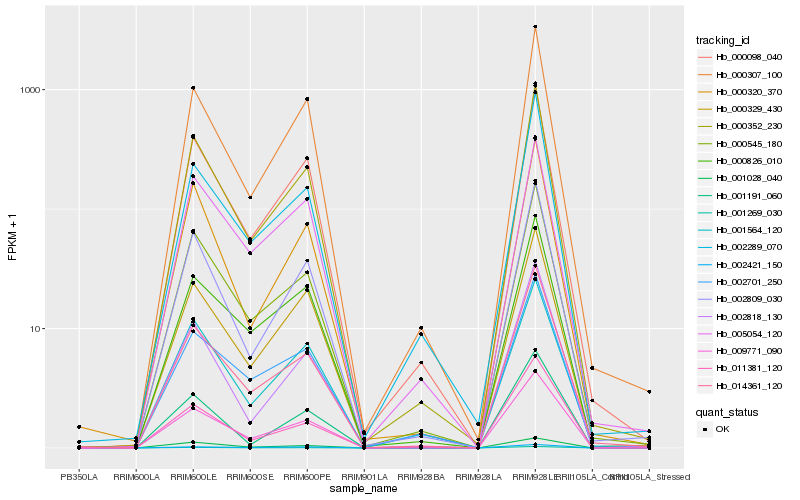
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009771_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000352_230 |
0.0346539342 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_000098_040 |
0.0500340647 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000329_430 |
0.054424666 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE10 [Jatropha curcas] |
| 5 |
Hb_000545_180 |
0.0563312435 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000307_100 |
0.0588254398 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_014361_120 |
0.059239468 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 8 |
Hb_000826_010 |
0.0614436797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002809_030 |
0.0628416317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001269_030 |
0.0670884702 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 11 |
Hb_002289_070 |
0.0812711331 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000320_370 |
0.0818247795 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002701_250 |
0.0819243697 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 14 |
Hb_001564_120 |
0.0838858954 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 15 |
Hb_011381_120 |
0.0861544755 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 16 |
Hb_002818_130 |
0.0863590008 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001028_040 |
0.0895697488 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 18 |
Hb_002421_150 |
0.0949111137 |
- |
- |
hypothetical protein PRUPE_ppa020511mg [Prunus persica] |
| 19 |
Hb_005054_120 |
0.0960461824 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001191_060 |
0.0965710657 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |