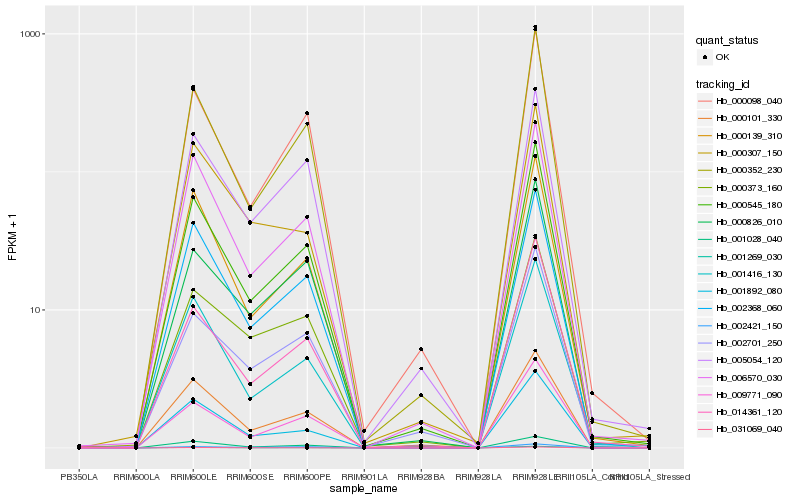
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_030 |
0.0 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 2 |
Hb_000545_180 |
0.0599543633 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001892_080 |
0.0635941021 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 4 |
Hb_009771_090 |
0.0670884702 |
- |
- |
- |
| 5 |
Hb_000826_010 |
0.0764682003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000373_160 |
0.0778152431 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 7 |
Hb_001028_040 |
0.0784217623 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 8 |
Hb_000352_230 |
0.078676479 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_002368_060 |
0.0796843608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002701_250 |
0.0812065797 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 11 |
Hb_014361_120 |
0.081718449 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 12 |
Hb_031069_040 |
0.0824043104 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_000101_330 |
0.0853081456 |
- |
- |
- |
| 14 |
Hb_000307_150 |
0.0908954475 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 15 |
Hb_006570_030 |
0.091989098 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005054_120 |
0.092339439 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001416_130 |
0.092714282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000098_040 |
0.0931729795 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000139_310 |
0.0943023066 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002421_150 |
0.094539393 |
- |
- |
hypothetical protein PRUPE_ppa020511mg [Prunus persica] |