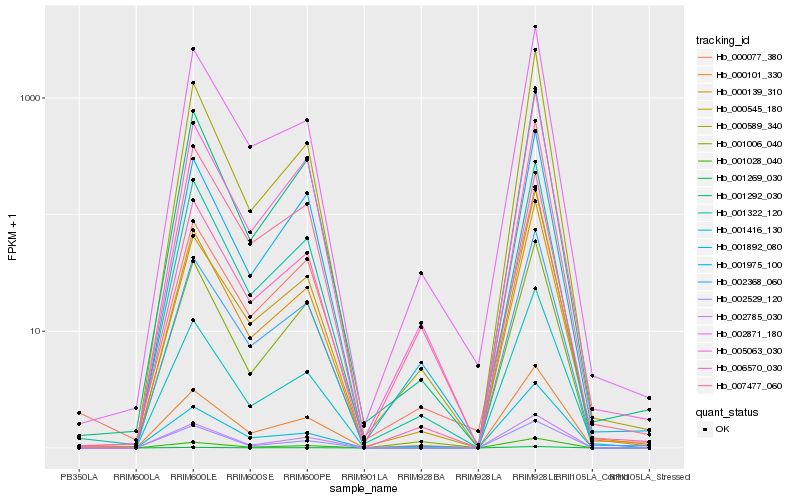
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001028_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 2 |
Hb_002368_060 |
0.0234356749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006570_030 |
0.0376882481 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000139_310 |
0.0414200706 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002785_030 |
0.0454079394 |
- |
- |
Polyketide cyclase/dehydrase and lipid transport superfamily protein [Theobroma cacao] |
| 6 |
Hb_001322_120 |
0.0578669328 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 7 |
Hb_001292_030 |
0.0581467712 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000101_330 |
0.0597598058 |
- |
- |
- |
| 9 |
Hb_001006_040 |
0.0602011399 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 10 |
Hb_001416_130 |
0.0618146641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000589_340 |
0.0632425377 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 12 |
Hb_005063_030 |
0.0652472028 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001892_080 |
0.067141591 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 14 |
Hb_001975_100 |
0.0699843759 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000545_180 |
0.0728825314 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_007477_060 |
0.0738641242 |
- |
- |
unknown [Medicago truncatula] |
| 17 |
Hb_000077_380 |
0.0748460488 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 18 |
Hb_002529_120 |
0.0761248127 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 19 |
Hb_001269_030 |
0.0784217623 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 20 |
Hb_002871_180 |
0.0796772181 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |