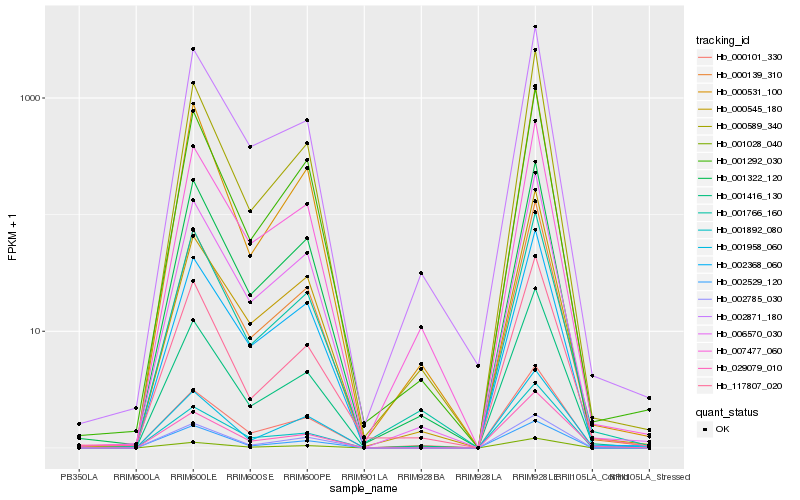
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_310 |
0.0 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001416_130 |
0.0328017908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006570_030 |
0.0330031002 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000589_340 |
0.0361078419 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_001028_040 |
0.0414200706 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 6 |
Hb_002368_060 |
0.0444322366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001292_030 |
0.0563800947 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001322_120 |
0.0574917066 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 9 |
Hb_002785_030 |
0.0577092456 |
- |
- |
Polyketide cyclase/dehydrase and lipid transport superfamily protein [Theobroma cacao] |
| 10 |
Hb_001892_080 |
0.058848212 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 11 |
Hb_029079_010 |
0.0650829917 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_002871_180 |
0.065915021 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000531_100 |
0.0673615828 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000101_330 |
0.0692226082 |
- |
- |
- |
| 15 |
Hb_007477_060 |
0.069738122 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_002529_120 |
0.0706310119 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 17 |
Hb_001766_160 |
0.0708203404 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 18 |
Hb_000545_180 |
0.0716549802 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001958_060 |
0.0719161256 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_117807_020 |
0.0729744549 |
- |
- |
methyltransferase, putative [Ricinus communis] |