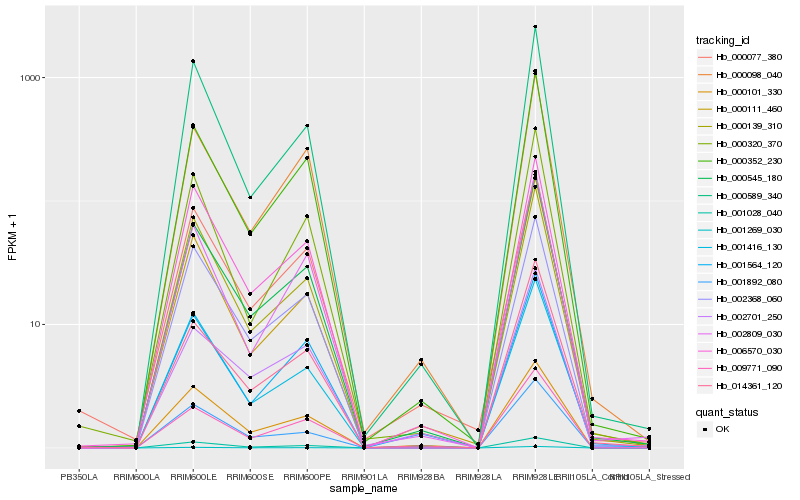
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000545_180 |
0.0 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000352_230 |
0.0386413049 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_014361_120 |
0.055158247 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 4 |
Hb_009771_090 |
0.0563312435 |
- |
- |
- |
| 5 |
Hb_000098_040 |
0.0583686049 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_001269_030 |
0.0599543633 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 7 |
Hb_001416_130 |
0.0603146045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002809_030 |
0.0683724853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000101_330 |
0.0688342475 |
- |
- |
- |
| 10 |
Hb_001892_080 |
0.0696101536 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 11 |
Hb_006570_030 |
0.0697392337 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000589_340 |
0.0704529984 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 13 |
Hb_000139_310 |
0.0716549802 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001564_120 |
0.0721522988 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 15 |
Hb_001028_040 |
0.0728825314 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 16 |
Hb_000320_370 |
0.0730209613 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000077_380 |
0.0741940748 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 18 |
Hb_002368_060 |
0.076341122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000111_460 |
0.0766984853 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 20 |
Hb_002701_250 |
0.080019057 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |