Hb_000307_100
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig307: 111091-111597 |
| Sequence |   |
Annotation
kegg
| ID | rcu:RCOM_1599580 |
|---|---|
| description | Plastocyanin A, chloroplast precursor, putative |
nr
| ID | XP_002510603.1 |
|---|---|
| description | Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
swissprot
| ID | P17340 |
|---|---|
| description | Plastocyanin, chloroplastic OS=Solanum lycopersicum GN=PETE PE=2 SV=1 |
trembl
| ID | B9R8G0 |
|---|---|
| description | Plastocyanin A, chloroplast, putative OS=Ricinus communis GN=RCOM_1599580 PE=4 SV=1 |
Gene Ontology
| ID | GO:0009535 |
|---|---|
| description | plastocyanin family protein |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_32417: 110999-112385 |
|---|---|
| cDNA (Sanger) (ID:Location) |
002_N20.ab1: 111031-111790 , 003_A01.ab1: 111031-111790 , 007_N10.ab1: 111019-111768 , 009_B24.ab1: 111031-111775 , 009_M13.ab1: 111031-111806 , 013_E14.ab1: 111031-111761 , 014_O18.ab1: 111031-111684 , 015_G20.ab1: 111063-111570 , 017_A01.ab1: 111041-111733 , 018_E12.ab1: 111031-111760 , 022_K17.ab1: 111019-111749 , 022_L12.ab1: 111031-111713 , 022_L18.ab1: 111027-111612 , 023_G13.ab1: 111019-111645 , 025_G17.ab1: 111031-111798 , 026_J16.ab1: 111044-111799 , 026_N18.ab1: 111031-111766 , 028_B17.ab1: 111031-111765 , 028_E20.ab1: 111058-111812 , 028_M11.ab1: 111031-111770 , 029_G06.ab1: 111031-111775 , 035_N23.ab1: 111031-111820 , 035_P12.ab1: 111031-111766 , 036_I21.ab1: 111031-111762 , 037_D09.ab1: 111031-111816 , 037_K17.ab1: 111031-111796 , 038_A14.ab1: 111019-111778 , 039_G05.ab1: 111031-111765 , 042_G04.ab1: 111031-111801 , 043_H20.ab1: 111019-111787 , 044_F06.ab1: 111489-111799 , 047_M12.ab1: 111031-111760 , 048_I18.ab1: 111031-111772 , 048_J17.ab1: 111031-111873 , 050_E08.ab1: 111031-111733 , 052_F14.ab1: 111031-111804 |
Similar expressed genes (Top20)
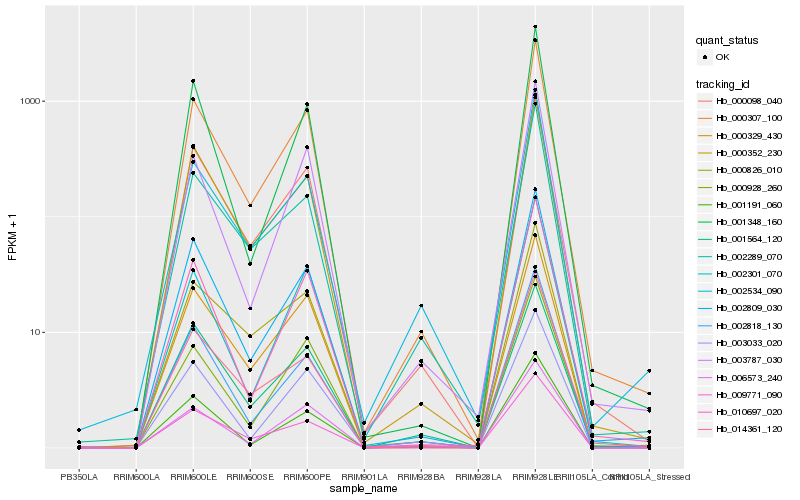
Gene co-expression network
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 0.0560785 | 124.094 | 1037.59 | 833.798 | 0 | 0.356966 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 3.67348 | 1.95837 | 0.175108 | 9.1579 | 3372.54 |

