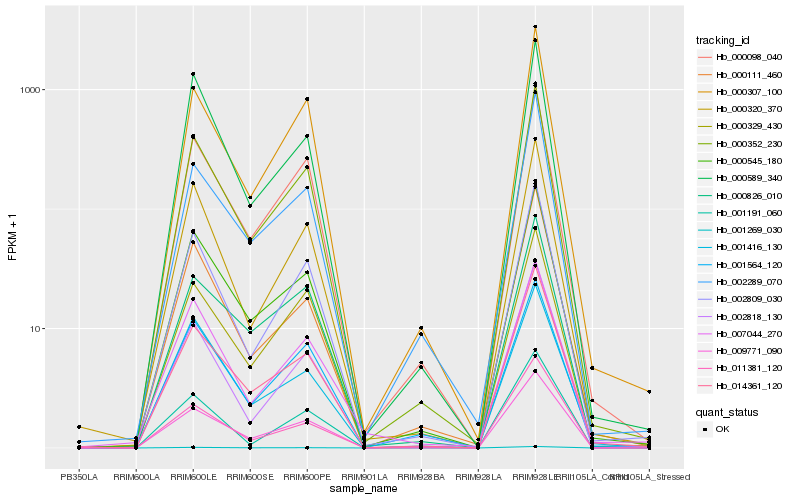
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_230 |
0.0 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_009771_090 |
0.0346539342 |
- |
- |
- |
| 3 |
Hb_000098_040 |
0.0381398662 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000545_180 |
0.0386413049 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_002809_030 |
0.0393292985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000307_100 |
0.0530867655 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_014361_120 |
0.0559162715 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 8 |
Hb_000320_370 |
0.0571291977 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000329_430 |
0.0664116146 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE10 [Jatropha curcas] |
| 10 |
Hb_001564_120 |
0.0669778698 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 11 |
Hb_002818_130 |
0.0743893954 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_011381_120 |
0.0769079226 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 13 |
Hb_002289_070 |
0.0786417165 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001269_030 |
0.078676479 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 15 |
Hb_000826_010 |
0.0792845808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000111_460 |
0.0795704868 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 17 |
Hb_001191_060 |
0.0799797179 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 18 |
Hb_000589_340 |
0.0801975656 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 19 |
Hb_001416_130 |
0.0814616977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007044_270 |
0.0833283698 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |