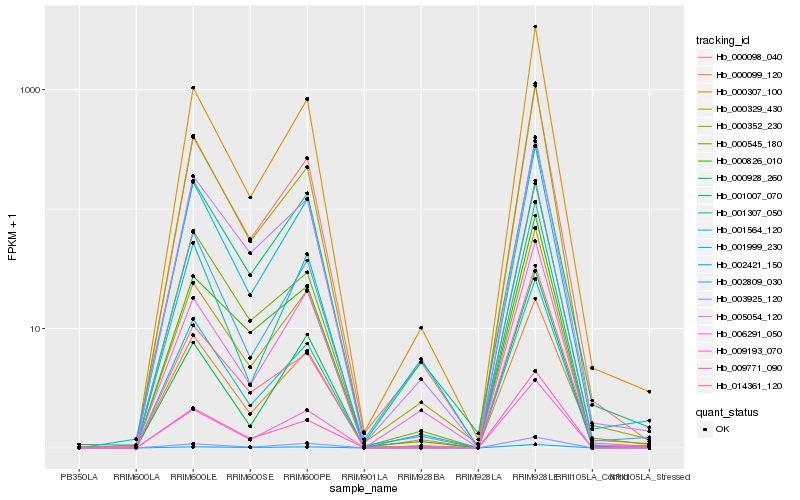
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_430 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE10 [Jatropha curcas] |
| 2 |
Hb_000307_100 |
0.0466831301 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000098_040 |
0.0496431086 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_009771_090 |
0.054424666 |
- |
- |
- |
| 5 |
Hb_003925_120 |
0.0569009485 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_000826_010 |
0.0638381668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000352_230 |
0.0664116146 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_009193_070 |
0.0696907 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 9 |
Hb_002809_030 |
0.072903062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001564_120 |
0.0796596761 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 11 |
Hb_001307_050 |
0.0825808905 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 12 |
Hb_001007_070 |
0.085316768 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 13 |
Hb_005054_120 |
0.0860628679 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002421_150 |
0.0861042199 |
- |
- |
hypothetical protein PRUPE_ppa020511mg [Prunus persica] |
| 15 |
Hb_006291_050 |
0.0905623783 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_000099_120 |
0.0909444721 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 17 |
Hb_000545_180 |
0.0913367311 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000928_260 |
0.092405576 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 19 |
Hb_014361_120 |
0.0925734375 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 20 |
Hb_001999_230 |
0.0925807841 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |