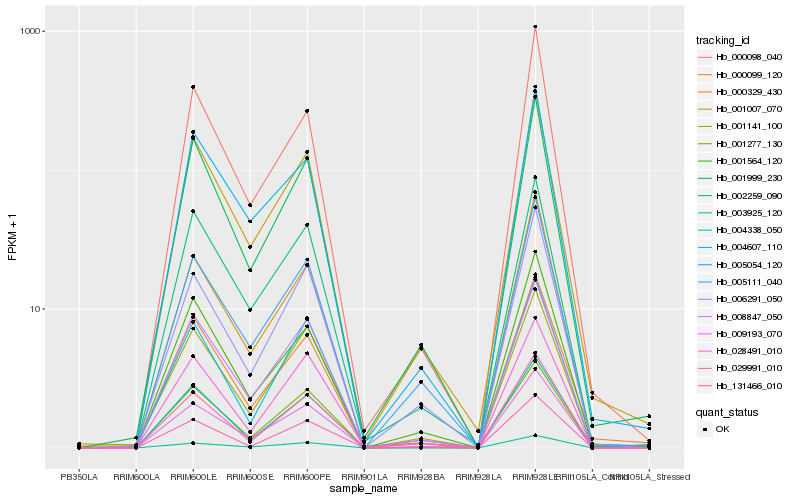
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009193_070 |
0.0 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 2 |
Hb_003925_120 |
0.0502442039 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_008847_050 |
0.052345018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001007_070 |
0.0536914055 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 5 |
Hb_028491_010 |
0.059393486 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 6 |
Hb_006291_050 |
0.0607397382 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_001999_230 |
0.0612886199 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 8 |
Hb_131466_010 |
0.0629268162 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 9 |
Hb_001141_100 |
0.0653050864 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 10 |
Hb_005111_040 |
0.06597917 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 11 |
Hb_004607_110 |
0.0669014211 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 12 |
Hb_000329_430 |
0.0696907 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE10 [Jatropha curcas] |
| 13 |
Hb_002259_090 |
0.0699680778 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 14 |
Hb_004338_050 |
0.0733876739 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 15 |
Hb_029991_010 |
0.0757017885 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 16 |
Hb_005054_120 |
0.0769770604 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001277_130 |
0.0781147726 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 18 |
Hb_000099_120 |
0.0783696253 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 19 |
Hb_000098_040 |
0.0788065762 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001564_120 |
0.0802596847 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |