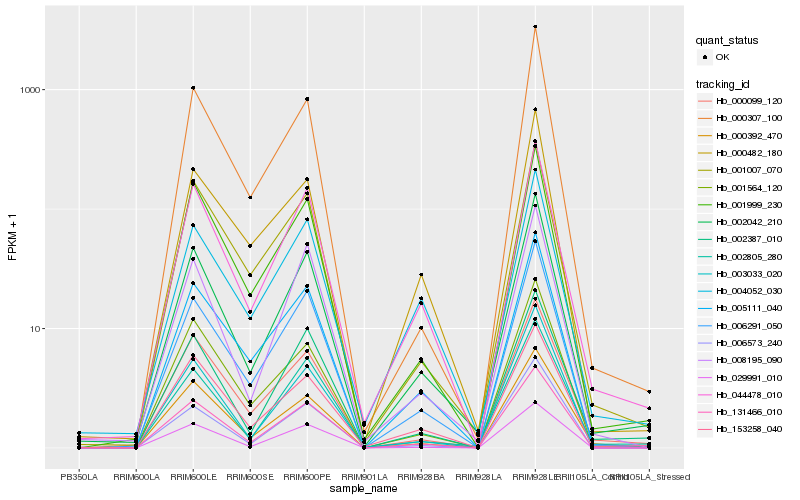
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_210 |
0.0 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 2 |
Hb_131466_010 |
0.0644364076 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 3 |
Hb_044478_010 |
0.0648881649 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 4 |
Hb_006291_050 |
0.0680069142 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_002805_280 |
0.0731043009 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 6 |
Hb_003033_020 |
0.0786322881 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 7 |
Hb_005111_040 |
0.0799002671 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 8 |
Hb_006573_240 |
0.0815723033 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_002387_010 |
0.0825353462 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 10 |
Hb_008195_090 |
0.0843341718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001999_230 |
0.0876648878 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 12 |
Hb_029991_010 |
0.0882051061 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 13 |
Hb_000392_470 |
0.0887276967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_153258_040 |
0.0891792026 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 15 |
Hb_001564_120 |
0.0896996099 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 16 |
Hb_000099_120 |
0.0901666926 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 17 |
Hb_000307_100 |
0.0919315015 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_001007_070 |
0.0928620838 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 19 |
Hb_000482_180 |
0.0934340029 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 20 |
Hb_004052_030 |
0.0947034289 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |