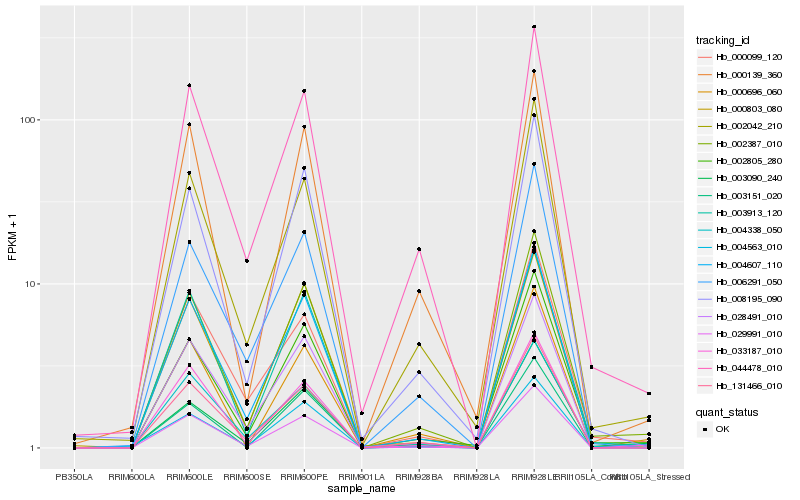
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002387_010 |
0.0 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002805_280 |
0.0462355998 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 3 |
Hb_004607_110 |
0.0706435551 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 4 |
Hb_008195_090 |
0.0741963777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_044478_010 |
0.0776139785 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 6 |
Hb_029991_010 |
0.0795571643 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 7 |
Hb_003913_120 |
0.0809577642 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 8 |
Hb_002042_210 |
0.0825353462 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003090_240 |
0.0840511788 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 10 |
Hb_131466_010 |
0.0854815894 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 11 |
Hb_033187_010 |
0.090077929 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 12 |
Hb_000139_360 |
0.0911206374 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 13 |
Hb_028491_010 |
0.0921862926 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 14 |
Hb_000696_060 |
0.0923600392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000803_080 |
0.0935715856 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 16 |
Hb_006291_050 |
0.0965688696 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_003151_020 |
0.0976145083 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 18 |
Hb_000099_120 |
0.0990754351 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 19 |
Hb_004338_050 |
0.0993365606 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 20 |
Hb_004563_010 |
0.1025364664 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |