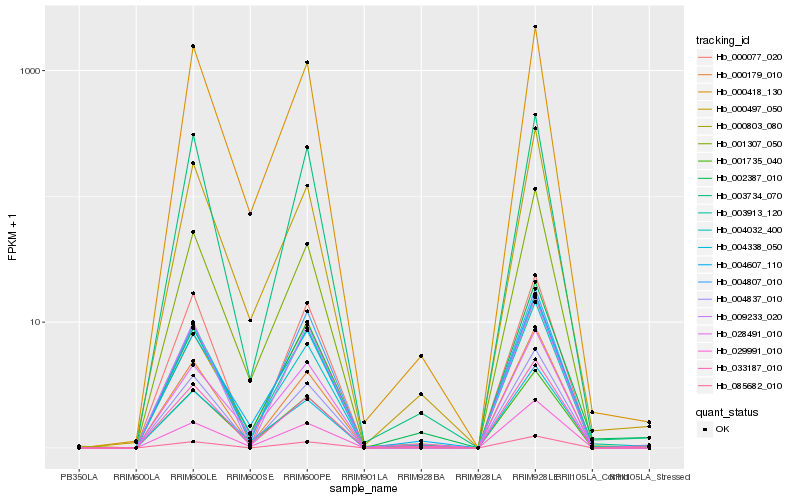
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003913_120 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 2 |
Hb_000803_080 |
0.0561295675 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 3 |
Hb_004837_010 |
0.0620042656 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 4 |
Hb_003734_070 |
0.0638230029 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 5 |
Hb_004607_110 |
0.06854991 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 6 |
Hb_004807_010 |
0.0713358985 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 7 |
Hb_000418_130 |
0.072282627 |
- |
- |
PREDICTED: photosystem I reaction center subunit psaK, chloroplastic [Jatropha curcas] |
| 8 |
Hb_029991_010 |
0.0723318237 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 9 |
Hb_001735_040 |
0.0735299917 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000179_010 |
0.0744113591 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 11 |
Hb_028491_010 |
0.0747995075 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 12 |
Hb_004338_050 |
0.0757324803 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 13 |
Hb_085682_010 |
0.0777837002 |
- |
- |
- |
| 14 |
Hb_004032_400 |
0.0781179568 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 15 |
Hb_033187_010 |
0.0792654722 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 16 |
Hb_001307_050 |
0.0800543298 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 17 |
Hb_009233_020 |
0.0807880756 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 18 |
Hb_002387_010 |
0.0809577642 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000497_050 |
0.0824280449 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000077_020 |
0.0832678066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |