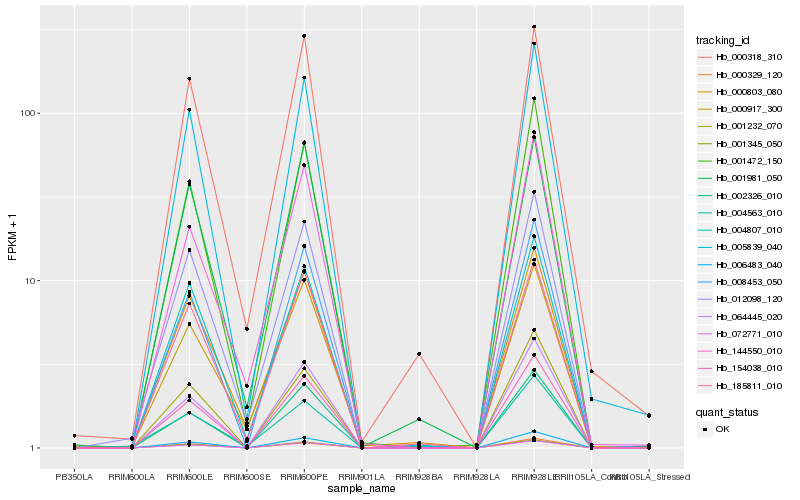
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012098_120 |
0.0 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 2 |
Hb_004807_010 |
0.0442665376 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 3 |
Hb_005839_040 |
0.0644187956 |
- |
- |
hypothetical protein POPTR_0019s08420g [Populus trichocarpa] |
| 4 |
Hb_185811_010 |
0.0683811786 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 5 |
Hb_000803_080 |
0.0690582329 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 6 |
Hb_001345_050 |
0.069192092 |
- |
- |
hypothetical protein POPTR_0014s14480g [Populus trichocarpa] |
| 7 |
Hb_072771_010 |
0.0692925569 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 8 |
Hb_154038_010 |
0.0709780202 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_001472_150 |
0.0724561504 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 10 |
Hb_006483_040 |
0.0739227365 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 11 |
Hb_000329_120 |
0.0753933003 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 12 |
Hb_008453_050 |
0.0783574185 |
- |
- |
PREDICTED: peroxidase 64 [Jatropha curcas] |
| 13 |
Hb_000917_300 |
0.0786806177 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 14 |
Hb_004563_010 |
0.0798405228 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001981_050 |
0.0823357309 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 16 |
Hb_000318_310 |
0.0842860206 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 17 |
Hb_001232_070 |
0.0853654771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002326_010 |
0.0856186228 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 19 |
Hb_064445_020 |
0.0869623447 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 20 |
Hb_144550_010 |
0.0877092591 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |