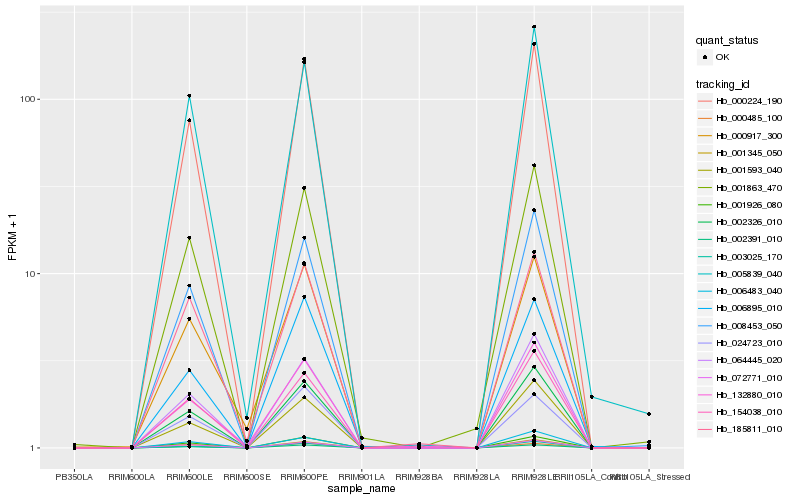
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000224_190 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_002326_010 |
0.023147224 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 3 |
Hb_001926_080 |
0.0262717595 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 4 |
Hb_002391_010 |
0.0342194468 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 5 |
Hb_072771_010 |
0.0354008391 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 6 |
Hb_001345_050 |
0.0361289516 |
- |
- |
hypothetical protein POPTR_0014s14480g [Populus trichocarpa] |
| 7 |
Hb_008453_050 |
0.0407805477 |
- |
- |
PREDICTED: peroxidase 64 [Jatropha curcas] |
| 8 |
Hb_154038_010 |
0.0461135225 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_064445_020 |
0.0486749441 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 10 |
Hb_000485_100 |
0.050776802 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 11 |
Hb_001593_040 |
0.0514623492 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_001863_470 |
0.0606967449 |
- |
- |
PREDICTED: uncharacterized protein LOC105630613 [Jatropha curcas] |
| 13 |
Hb_006483_040 |
0.0635888142 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 14 |
Hb_003025_170 |
0.0705843908 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |
| 15 |
Hb_132880_010 |
0.0732687812 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 16 |
Hb_006895_010 |
0.0750707452 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 17 |
Hb_185811_010 |
0.0755234993 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_005839_040 |
0.0765704157 |
- |
- |
hypothetical protein POPTR_0019s08420g [Populus trichocarpa] |
| 19 |
Hb_024723_010 |
0.078727032 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 20 |
Hb_000917_300 |
0.0801253545 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |