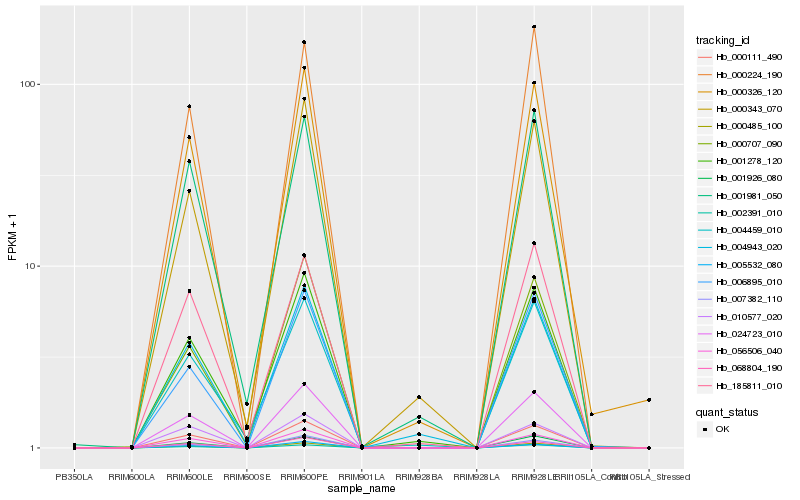
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024723_010 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 2 |
Hb_000111_490 |
0.0141060908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000485_100 |
0.035476644 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 4 |
Hb_002391_010 |
0.04476423 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 5 |
Hb_056506_040 |
0.0471408918 |
- |
- |
- |
| 6 |
Hb_001926_080 |
0.0538937757 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 7 |
Hb_000326_120 |
0.0609705692 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_068804_190 |
0.0660375346 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_005532_080 |
0.0701245791 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 10 |
Hb_000343_070 |
0.0751477547 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 11 |
Hb_001278_120 |
0.0759554293 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 12 |
Hb_007382_110 |
0.0772774174 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 13 |
Hb_000224_190 |
0.078727032 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000707_090 |
0.082020461 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 15 |
Hb_006895_010 |
0.0835460449 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 16 |
Hb_010577_020 |
0.0835759693 |
- |
- |
- |
| 17 |
Hb_001981_050 |
0.0855491956 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 18 |
Hb_004943_020 |
0.0865598454 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_004459_010 |
0.0883459712 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 20 |
Hb_185811_010 |
0.0917422682 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |