| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
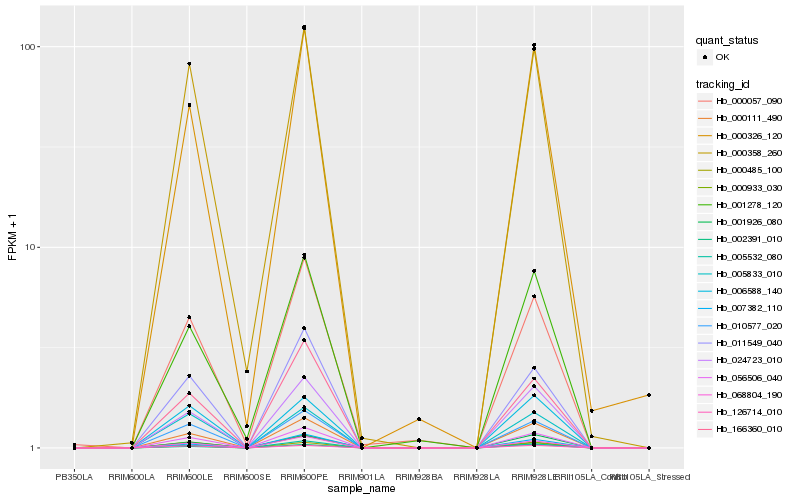
Hb_056506_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000111_490 |
0.0338942651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010577_020 |
0.0385876526 |
- |
- |
- |
| 4 |
Hb_068804_190 |
0.0391405009 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_024723_010 |
0.0471408918 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 6 |
Hb_005532_080 |
0.0565829411 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 7 |
Hb_011549_040 |
0.0592508386 |
- |
- |
- |
| 8 |
Hb_166360_010 |
0.0706358126 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 9 |
Hb_000358_260 |
0.0728343563 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Vitis vinifera] |
| 10 |
Hb_000326_120 |
0.077922319 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000485_100 |
0.0821328016 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 12 |
Hb_000057_090 |
0.085601261 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 13 |
Hb_002391_010 |
0.0859974503 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 14 |
Hb_005833_010 |
0.086878254 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 15 |
Hb_006588_140 |
0.0897555366 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_126714_010 |
0.0916505896 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 17 |
Hb_001926_080 |
0.0925987483 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 18 |
Hb_007382_110 |
0.0928892734 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 19 |
Hb_000933_030 |
0.0930858827 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001278_120 |
0.0948831957 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |