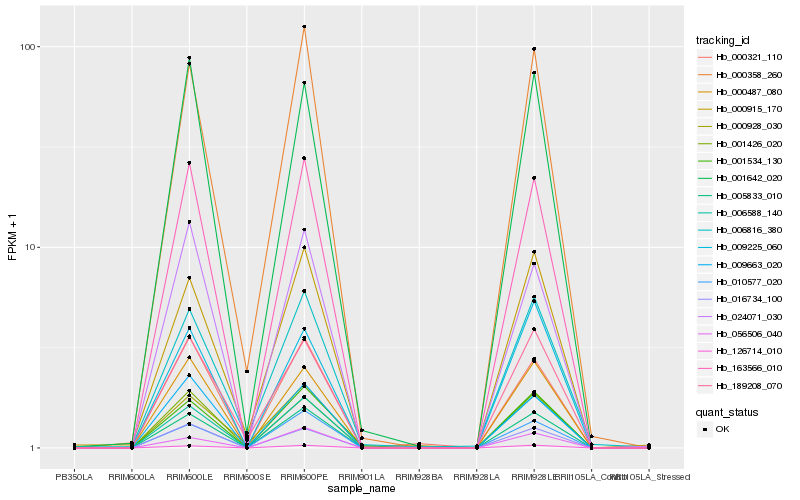
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005833_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 2 |
Hb_126714_010 |
0.0391539792 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 3 |
Hb_163566_010 |
0.0393871822 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Amborella trichopoda] |
| 4 |
Hb_006588_140 |
0.0445981929 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_010577_020 |
0.0577336048 |
- |
- |
- |
| 6 |
Hb_000358_260 |
0.0650373144 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Vitis vinifera] |
| 7 |
Hb_001426_020 |
0.0665058453 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 8 |
Hb_000487_080 |
0.0743554263 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 9 |
Hb_001534_130 |
0.0760111886 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 10 |
Hb_016734_100 |
0.0805818902 |
- |
- |
- |
| 11 |
Hb_056506_040 |
0.086878254 |
- |
- |
- |
| 12 |
Hb_009663_020 |
0.0909123644 |
- |
- |
- |
| 13 |
Hb_000915_170 |
0.0931077964 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 14 |
Hb_000321_110 |
0.0936707618 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 15 |
Hb_001642_020 |
0.0937698155 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 16 |
Hb_000928_030 |
0.0976306586 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 17 |
Hb_189208_070 |
0.0989824409 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |
| 18 |
Hb_024071_030 |
0.0996645528 |
- |
- |
hypothetical protein CICLE_v10025479mg [Citrus clementina] |
| 19 |
Hb_009225_060 |
0.1020859835 |
- |
- |
PREDICTED: myosin heavy chain kinase B-like [Populus euphratica] |
| 20 |
Hb_006816_380 |
0.102146275 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |