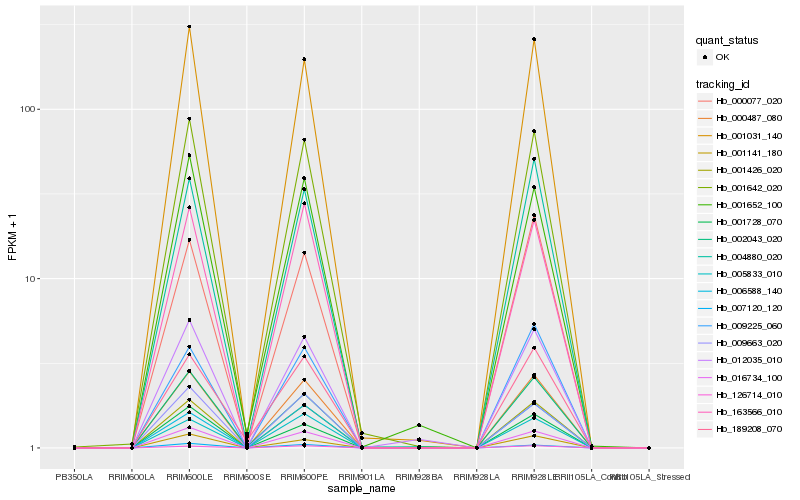
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001426_020 |
0.0 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 2 |
Hb_000487_080 |
0.0256632942 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 3 |
Hb_016734_100 |
0.0276807704 |
- |
- |
- |
| 4 |
Hb_001642_020 |
0.0417921634 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 5 |
Hb_001031_140 |
0.0518704948 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 6 |
Hb_163566_010 |
0.0581310322 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Amborella trichopoda] |
| 7 |
Hb_005833_010 |
0.0665058453 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001141_180 |
0.0680009157 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 9 |
Hb_126714_010 |
0.0680826003 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 10 |
Hb_004880_020 |
0.0699850331 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Jatropha curcas] |
| 11 |
Hb_006588_140 |
0.0737505847 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_012035_010 |
0.0744419128 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 13 |
Hb_009663_020 |
0.0748288484 |
- |
- |
- |
| 14 |
Hb_007120_120 |
0.0756717535 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_009225_060 |
0.0823847392 |
- |
- |
PREDICTED: myosin heavy chain kinase B-like [Populus euphratica] |
| 16 |
Hb_002043_020 |
0.0844326763 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 17 |
Hb_001652_100 |
0.0845026081 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 18 |
Hb_189208_070 |
0.0853589603 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000077_020 |
0.0893635686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001728_070 |
0.0896253092 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |