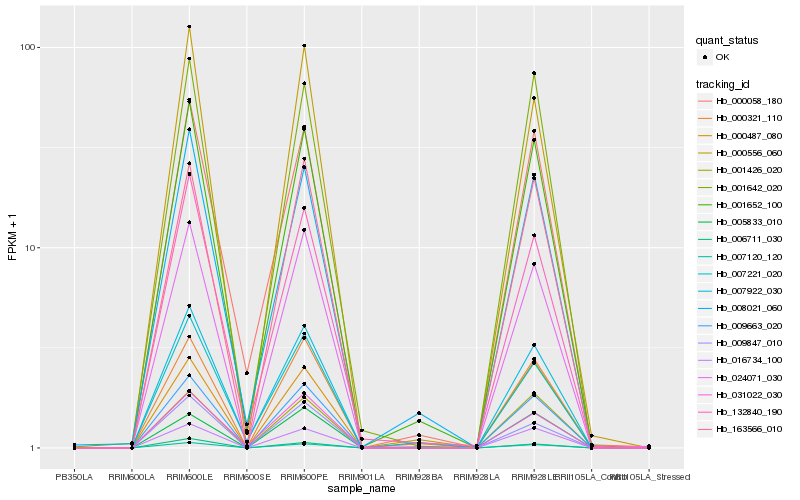
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009663_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_007120_120 |
0.0189763172 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_024071_030 |
0.0353126277 |
- |
- |
hypothetical protein CICLE_v10025479mg [Citrus clementina] |
| 4 |
Hb_163566_010 |
0.0520014485 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Amborella trichopoda] |
| 5 |
Hb_016734_100 |
0.0530609082 |
- |
- |
- |
| 6 |
Hb_001652_100 |
0.057324799 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 7 |
Hb_000321_110 |
0.0668773573 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 8 |
Hb_000556_060 |
0.0674648408 |
- |
- |
hypothetical protein OsJ_28181 [Oryza sativa Japonica Group] |
| 9 |
Hb_001642_020 |
0.074445719 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 10 |
Hb_001426_020 |
0.0748288484 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 11 |
Hb_000487_080 |
0.077821635 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 12 |
Hb_031022_030 |
0.0795262284 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7-like [Jatropha curcas] |
| 13 |
Hb_009847_010 |
0.0797107841 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 14 |
Hb_132840_190 |
0.0858634872 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000058_180 |
0.0863782766 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 16 |
Hb_008021_060 |
0.0880164193 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 17 |
Hb_007922_030 |
0.0887122329 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 18 |
Hb_006711_030 |
0.0898655489 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 19 |
Hb_007221_020 |
0.0906598725 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 20 |
Hb_005833_010 |
0.0909123644 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |