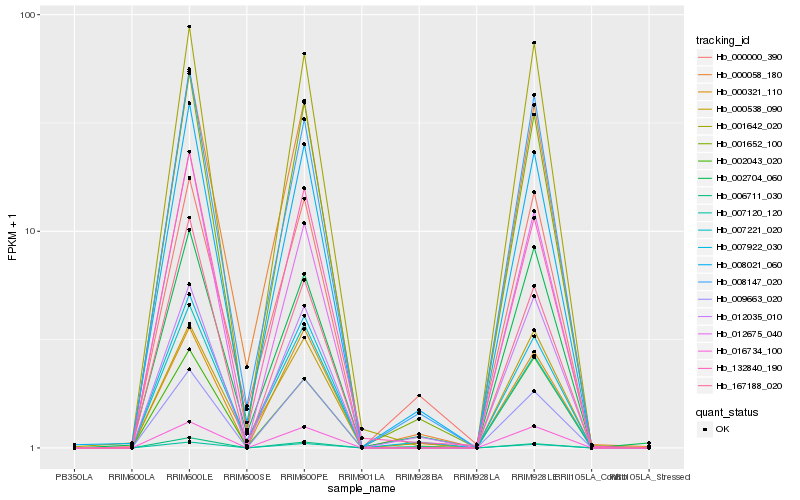
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008021_060 |
0.0 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_001652_100 |
0.0423978376 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 3 |
Hb_007922_030 |
0.0488928226 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 4 |
Hb_000058_180 |
0.0645155063 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 5 |
Hb_008147_020 |
0.0651403601 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 6 |
Hb_132840_190 |
0.0692056045 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_007120_120 |
0.0788927161 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_007221_020 |
0.0800823945 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_002704_060 |
0.0849084622 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_012035_010 |
0.0853033659 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 11 |
Hb_009663_020 |
0.0880164193 |
- |
- |
- |
| 12 |
Hb_001642_020 |
0.0888729204 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 13 |
Hb_000538_090 |
0.0900631193 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 14 |
Hb_012675_040 |
0.0906902989 |
- |
- |
PREDICTED: uncharacterized protein LOC105647773 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002043_020 |
0.0916282048 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 16 |
Hb_000321_110 |
0.0919910212 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 17 |
Hb_000000_390 |
0.0929450406 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 18 |
Hb_016734_100 |
0.0932590768 |
- |
- |
- |
| 19 |
Hb_006711_030 |
0.0946551018 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 20 |
Hb_167188_020 |
0.0963759164 |
- |
- |
PREDICTED: uncharacterized protein At1g05835 [Jatropha curcas] |