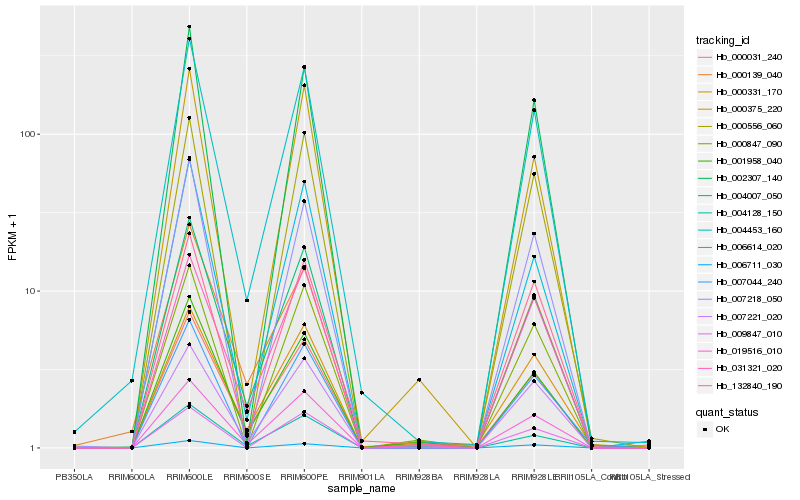
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004453_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004007_050 |
0.0547955734 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 3 |
Hb_007221_020 |
0.0694203916 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_132840_190 |
0.0722058904 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000031_240 |
0.0787000921 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_004128_150 |
0.08443422 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 7 |
Hb_002307_140 |
0.0865153747 |
transcription factor |
TF Family: AUX/IAA |
aux/IAA family protein [Populus trichocarpa] |
| 8 |
Hb_031321_020 |
0.0869346412 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 9 |
Hb_000331_170 |
0.0884571427 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 10 |
Hb_006614_020 |
0.0894511757 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 11 |
Hb_019516_010 |
0.0901362369 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 12 |
Hb_001958_040 |
0.0901559804 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 13 |
Hb_007044_240 |
0.0912172863 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007218_050 |
0.0916722205 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000139_040 |
0.0926776 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 16 |
Hb_000556_060 |
0.0940000094 |
- |
- |
hypothetical protein OsJ_28181 [Oryza sativa Japonica Group] |
| 17 |
Hb_000375_220 |
0.0946689596 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 18 |
Hb_000847_090 |
0.094980006 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006711_030 |
0.0951144705 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 20 |
Hb_009847_010 |
0.0957135767 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |