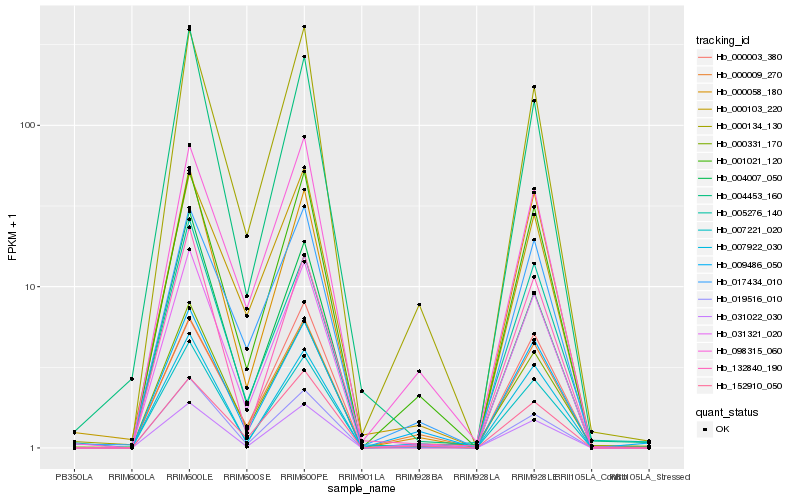
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031321_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 2 |
Hb_007221_020 |
0.0376662669 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_031022_030 |
0.055268059 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7-like [Jatropha curcas] |
| 4 |
Hb_001021_120 |
0.0577439504 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 5 |
Hb_007922_030 |
0.0646283452 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 6 |
Hb_000134_130 |
0.0729625438 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 7 |
Hb_000331_170 |
0.073359001 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 8 |
Hb_019516_010 |
0.0766615076 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 9 |
Hb_017434_010 |
0.078131959 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 10 |
Hb_000058_180 |
0.0805050445 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 11 |
Hb_152910_050 |
0.0810671466 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000009_270 |
0.0817196119 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 13 |
Hb_132840_190 |
0.0843281638 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_004453_160 |
0.0869346412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_098315_060 |
0.0882891498 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 16 |
Hb_004007_050 |
0.0895767895 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 17 |
Hb_000103_220 |
0.0912244349 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 18 |
Hb_005276_140 |
0.0945041526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009486_050 |
0.0955143771 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 20 |
Hb_000003_380 |
0.096419078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |