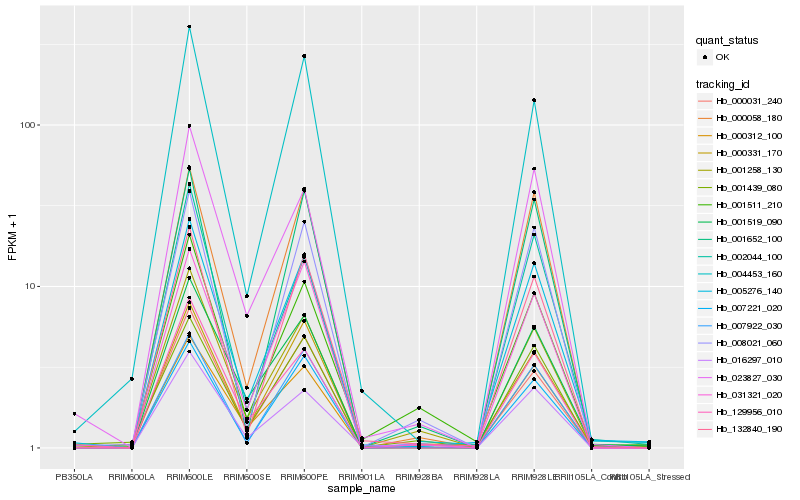
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005276_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000331_170 |
0.0797418437 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |
| 3 |
Hb_132840_190 |
0.0830343157 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000058_180 |
0.083228283 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 5 |
Hb_000312_100 |
0.0872668464 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_001519_090 |
0.0889531685 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 7 |
Hb_007221_020 |
0.0924231854 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_031321_020 |
0.0945041526 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 9 |
Hb_023827_030 |
0.0948994798 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_008021_060 |
0.0971382912 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_007922_030 |
0.0983119755 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 12 |
Hb_129956_010 |
0.099861168 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 13 |
Hb_004453_160 |
0.1009992604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_016297_010 |
0.1023102504 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 15 |
Hb_001439_080 |
0.1036038268 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 16 |
Hb_000031_240 |
0.1044628169 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_001652_100 |
0.1068670638 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 18 |
Hb_002044_100 |
0.1076150618 |
- |
- |
PREDICTED: uncharacterized protein LOC105643555 [Jatropha curcas] |
| 19 |
Hb_001258_130 |
0.1078556868 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 20 |
Hb_001511_210 |
0.1096515039 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |