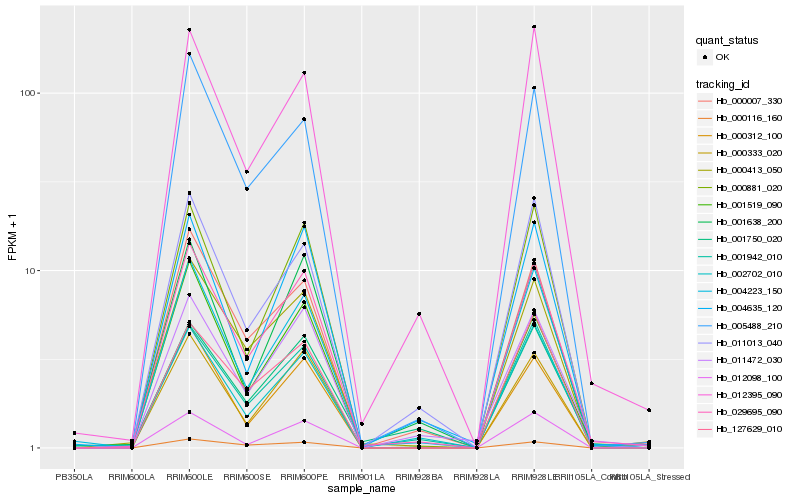
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029695_090 |
0.0 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 2 |
Hb_000413_050 |
0.0673432925 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004223_150 |
0.0762706271 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 4 |
Hb_000007_330 |
0.0826365876 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 5 |
Hb_011472_030 |
0.083947962 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 6 |
Hb_012395_090 |
0.0886851899 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000881_020 |
0.0895030144 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 8 |
Hb_005488_210 |
0.0897135911 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 9 |
Hb_000312_100 |
0.0920598967 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 10 |
Hb_000333_020 |
0.0929024756 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 11 |
Hb_004635_120 |
0.0962664588 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 12 |
Hb_001519_090 |
0.097103602 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 13 |
Hb_002702_010 |
0.0980619631 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 14 |
Hb_001942_010 |
0.0984476861 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 15 |
Hb_011013_040 |
0.1007908732 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 16 |
Hb_000116_160 |
0.1031711976 |
transcription factor |
TF Family: RWP-RK |
hypothetical protein JCGZ_22391 [Jatropha curcas] |
| 17 |
Hb_001638_200 |
0.1035357185 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 18 |
Hb_001750_020 |
0.1054725765 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_127629_010 |
0.1068218269 |
- |
- |
hypothetical protein CICLE_v10005216mg [Citrus clementina] |
| 20 |
Hb_012098_100 |
0.1088498414 |
- |
- |
zinc finger protein, putative [Ricinus communis] |