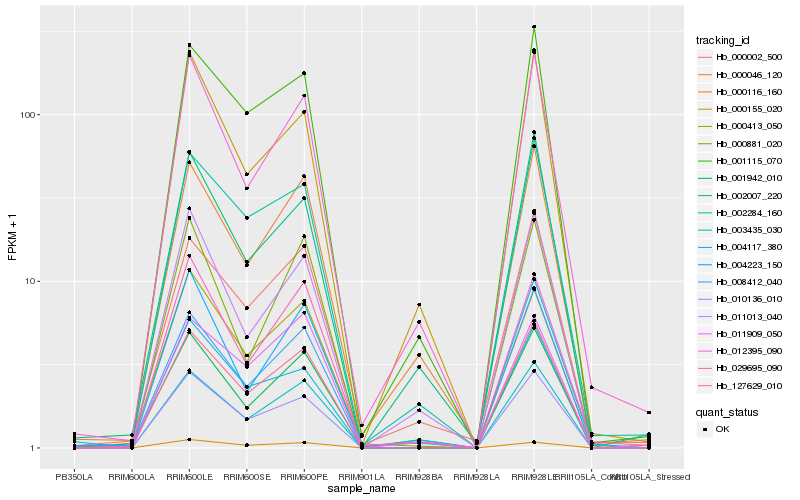
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127629_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10005216mg [Citrus clementina] |
| 2 |
Hb_008412_040 |
0.062641214 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_001115_070 |
0.0699144995 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 4 |
Hb_002284_160 |
0.0817658359 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_004117_380 |
0.0895903653 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 6 |
Hb_000002_500 |
0.0939043469 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 7 |
Hb_010136_010 |
0.0964583056 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 8 |
Hb_001942_010 |
0.0972186776 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 9 |
Hb_012395_090 |
0.0986623674 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003435_030 |
0.0993567825 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_000413_050 |
0.0999955627 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000881_020 |
0.1033139891 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 13 |
Hb_000046_120 |
0.1041210679 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_000116_160 |
0.1059463768 |
transcription factor |
TF Family: RWP-RK |
hypothetical protein JCGZ_22391 [Jatropha curcas] |
| 15 |
Hb_029695_090 |
0.1068218269 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 16 |
Hb_011013_040 |
0.1098010412 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 17 |
Hb_000155_020 |
0.1099434371 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_011909_050 |
0.1104132104 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 19 |
Hb_004223_150 |
0.1143356437 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 20 |
Hb_002007_220 |
0.1150234691 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |