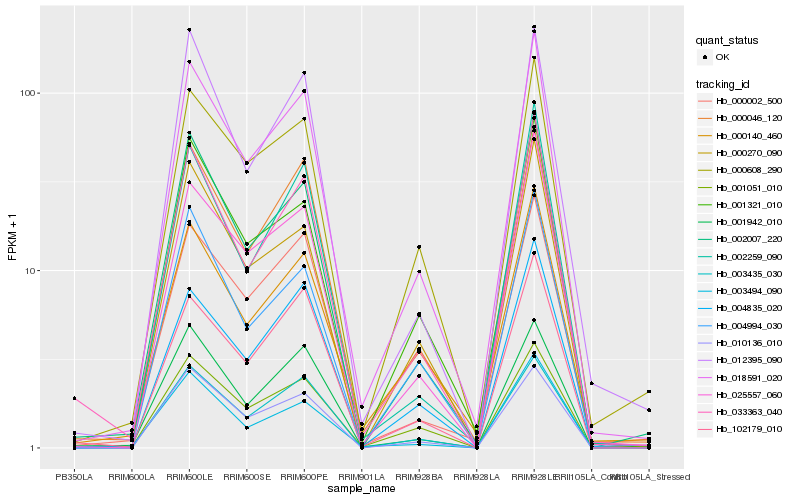
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018591_020 |
0.0 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_002007_220 |
0.0596845143 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000046_120 |
0.0609950362 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_033363_040 |
0.0651707617 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 5 |
Hb_025557_060 |
0.0682464756 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 6 |
Hb_000608_290 |
0.0696405181 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 7 |
Hb_000002_500 |
0.0733437523 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003435_030 |
0.0751132865 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_001321_010 |
0.0755314242 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 10 |
Hb_001051_010 |
0.0790578654 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 11 |
Hb_010136_010 |
0.0798079075 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 12 |
Hb_004835_020 |
0.0799476699 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_002259_090 |
0.0807209977 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 14 |
Hb_000270_090 |
0.0808435064 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 15 |
Hb_003494_090 |
0.0816026134 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 16 |
Hb_000140_460 |
0.0824625286 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 17 |
Hb_004994_030 |
0.0844255192 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 18 |
Hb_102179_010 |
0.0847434516 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_001942_010 |
0.087484882 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 20 |
Hb_012395_090 |
0.0884508134 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |