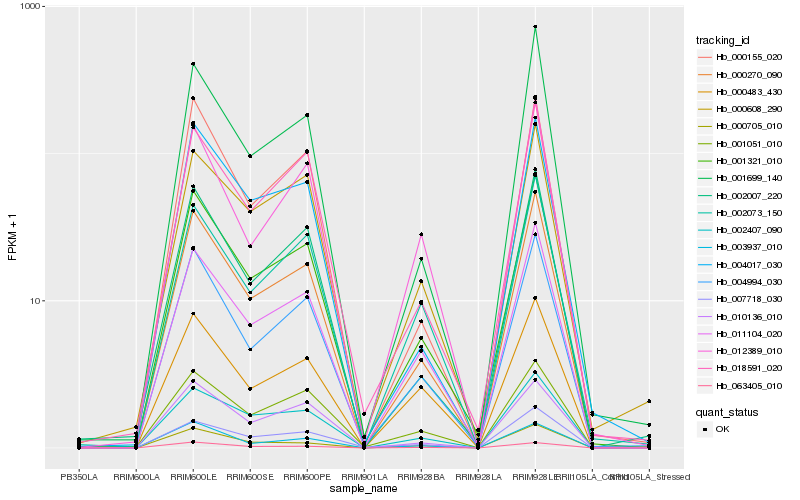
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001321_010 |
0.0 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 2 |
Hb_000270_090 |
0.0354122285 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 3 |
Hb_004994_030 |
0.0469457218 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 4 |
Hb_011104_020 |
0.0589406697 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 5 |
Hb_002007_220 |
0.073958946 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_018591_020 |
0.0755314242 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002407_090 |
0.0769744658 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 8 |
Hb_003937_010 |
0.0805492977 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 9 |
Hb_000155_020 |
0.0823952294 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001699_140 |
0.0835887015 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004017_030 |
0.0851145309 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 12 |
Hb_002073_150 |
0.0864275822 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 13 |
Hb_000608_290 |
0.0873619088 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 14 |
Hb_007718_030 |
0.0876826453 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 15 |
Hb_063405_010 |
0.0899981232 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 16 |
Hb_010136_010 |
0.0919826841 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 17 |
Hb_000483_430 |
0.0924719443 |
- |
- |
- |
| 18 |
Hb_001051_010 |
0.0925328182 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 19 |
Hb_012389_010 |
0.0933027665 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 20 |
Hb_000705_010 |
0.0936706961 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |