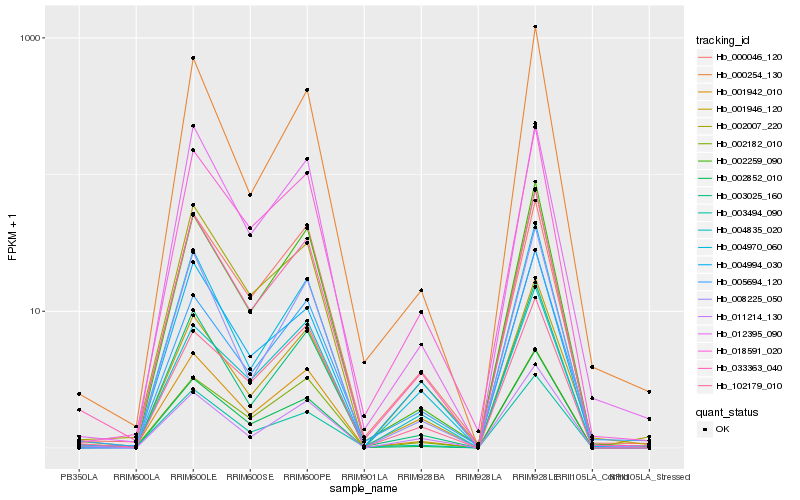
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033363_040 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 2 |
Hb_018591_020 |
0.0651707617 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_002007_220 |
0.0654860747 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003025_160 |
0.0659806272 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 5 |
Hb_004970_060 |
0.0695266237 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 6 |
Hb_002259_090 |
0.0776968183 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 7 |
Hb_003494_090 |
0.0815083404 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_001946_120 |
0.0816191641 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_011214_130 |
0.0818102714 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 10 |
Hb_002182_010 |
0.0818200372 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Jatropha curcas] |
| 11 |
Hb_000046_120 |
0.0821886841 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_001942_010 |
0.0851949775 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 13 |
Hb_002852_010 |
0.0873003709 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_000254_130 |
0.0878420169 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_005694_120 |
0.0881940089 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 16 |
Hb_012395_090 |
0.0909287577 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 17 |
Hb_102179_010 |
0.0917312711 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_008225_050 |
0.0936690747 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 19 |
Hb_004994_030 |
0.09458521 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 20 |
Hb_004835_020 |
0.0957518456 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |