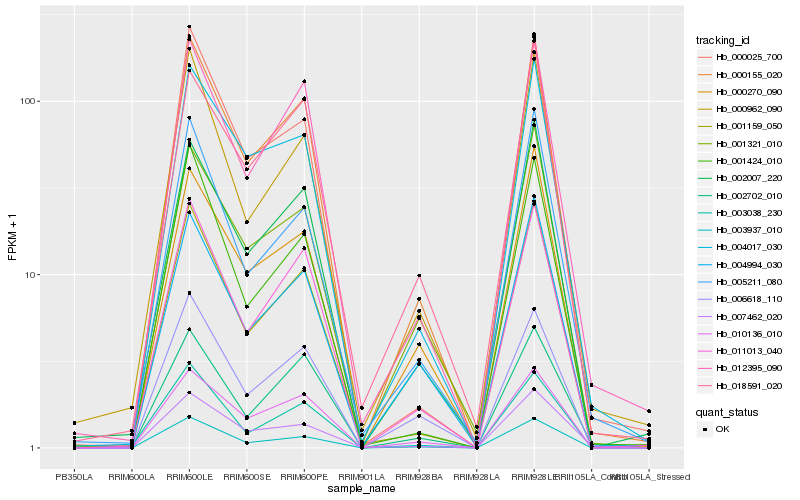
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000155_020 |
0.0 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_011013_040 |
0.0484948203 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 3 |
Hb_001159_050 |
0.0539240331 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 4 |
Hb_012395_090 |
0.0594105817 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002007_220 |
0.0602895916 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_010136_010 |
0.0605157174 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 7 |
Hb_004017_030 |
0.063451278 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 8 |
Hb_003937_010 |
0.0739363555 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 9 |
Hb_000270_090 |
0.0763490363 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 10 |
Hb_000025_700 |
0.0801859884 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 11 |
Hb_004994_030 |
0.0814011554 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 12 |
Hb_000962_090 |
0.0814974859 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 13 |
Hb_001321_010 |
0.0823952294 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 14 |
Hb_002702_010 |
0.0844329776 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 15 |
Hb_007462_020 |
0.0857227404 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_006618_110 |
0.0867741959 |
- |
- |
- |
| 17 |
Hb_005211_080 |
0.0877978606 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 18 |
Hb_003038_230 |
0.0888380256 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 19 |
Hb_001424_010 |
0.0890282901 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_018591_020 |
0.0890790645 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |