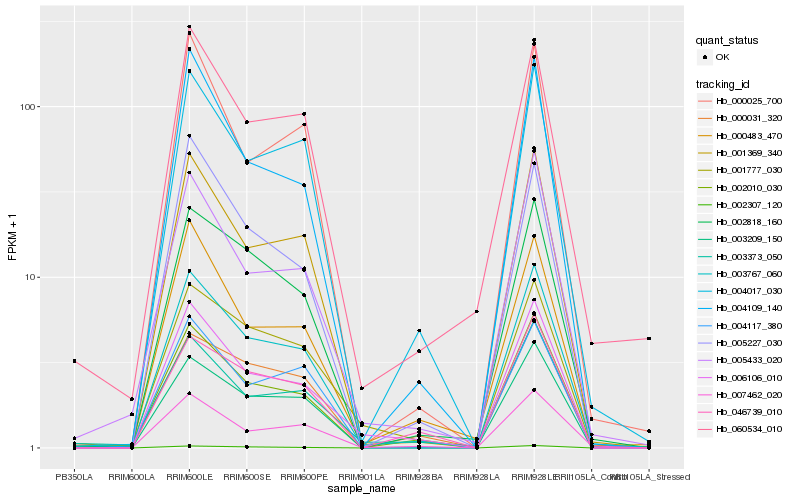
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003767_060 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 2 |
Hb_001369_340 |
0.0570309188 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 3 |
Hb_002010_030 |
0.0615418933 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_004017_030 |
0.0665174183 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 5 |
Hb_003209_150 |
0.0776260238 |
- |
- |
CLE9, putative [Ricinus communis] |
| 6 |
Hb_007462_020 |
0.0783634189 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 7 |
Hb_002818_160 |
0.0802359642 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 8 |
Hb_006106_010 |
0.081521056 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002307_120 |
0.0909080734 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 10 |
Hb_004109_140 |
0.0913149426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005433_020 |
0.0962086533 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 12 |
Hb_000025_700 |
0.0990517202 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 13 |
Hb_004117_380 |
0.0996113063 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 14 |
Hb_000031_320 |
0.0999967063 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001777_030 |
0.1011934433 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 16 |
Hb_000483_470 |
0.1013590643 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_046739_010 |
0.1019679951 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003373_050 |
0.1043833091 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 19 |
Hb_060534_010 |
0.1074055044 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 20 |
Hb_005227_030 |
0.1092800855 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |