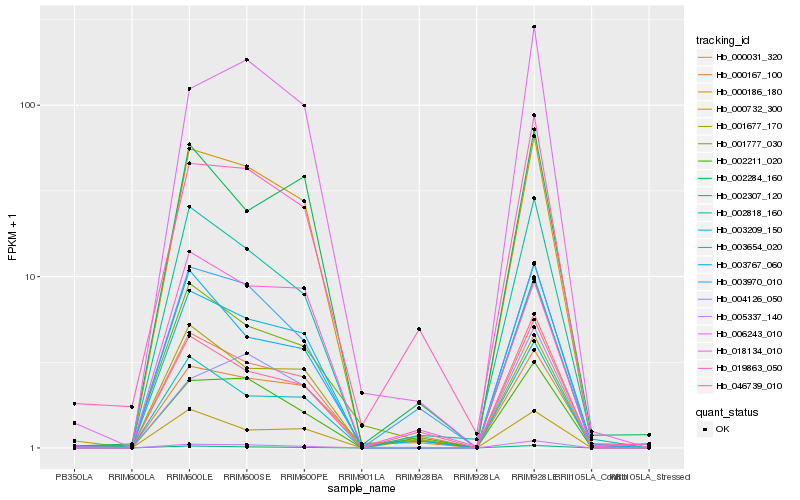
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 2 |
Hb_003654_020 |
0.0549248152 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_002818_160 |
0.0946235613 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 4 |
Hb_000031_320 |
0.0977074488 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000167_100 |
0.1057958968 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002307_120 |
0.1077741442 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 7 |
Hb_001677_170 |
0.1119870054 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 8 |
Hb_003970_010 |
0.114523412 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_001777_030 |
0.1148356193 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 10 |
Hb_005337_140 |
0.1152943684 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 11 |
Hb_003209_150 |
0.1155505459 |
- |
- |
CLE9, putative [Ricinus communis] |
| 12 |
Hb_006243_010 |
0.1165093338 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 13 |
Hb_000732_300 |
0.1229112003 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002211_020 |
0.1261864774 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 15 |
Hb_002284_160 |
0.1322613697 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_018134_010 |
0.136552845 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 17 |
Hb_046739_010 |
0.137272196 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004126_050 |
0.1376820638 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 19 |
Hb_003767_060 |
0.1379052519 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 20 |
Hb_019863_050 |
0.1392950256 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |