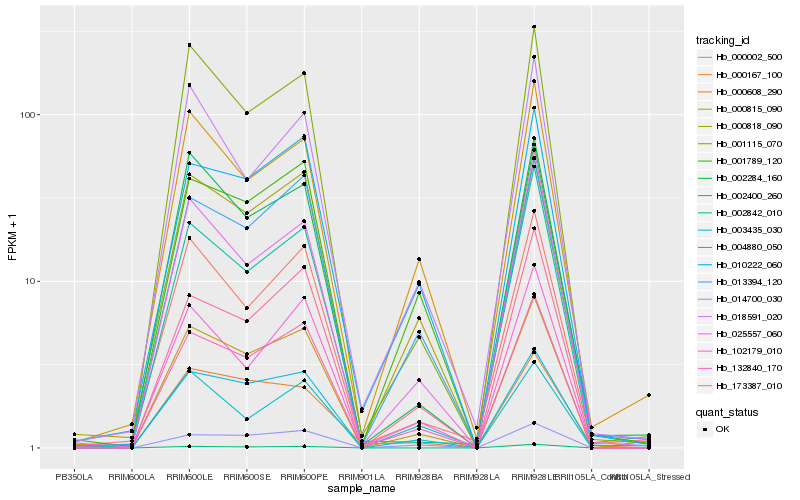
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 2 |
Hb_132840_170 |
0.0761362858 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 3 |
Hb_013394_120 |
0.0935597194 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 4 |
Hb_002284_160 |
0.0939620868 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000002_500 |
0.0948617808 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001115_070 |
0.0954148212 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 7 |
Hb_000818_090 |
0.0968554404 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 8 |
Hb_001789_120 |
0.1078674042 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000608_290 |
0.1081526994 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 10 |
Hb_010222_060 |
0.1097770936 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_102179_010 |
0.1152683827 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_014700_030 |
0.1156059914 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 13 |
Hb_173387_010 |
0.1161634317 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 14 |
Hb_004880_050 |
0.1166405667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_025557_060 |
0.1195978332 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_002842_010 |
0.1220123226 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 17 |
Hb_003435_030 |
0.1246112757 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002400_260 |
0.1262042155 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_018591_020 |
0.1264641938 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000167_100 |
0.1269929074 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |