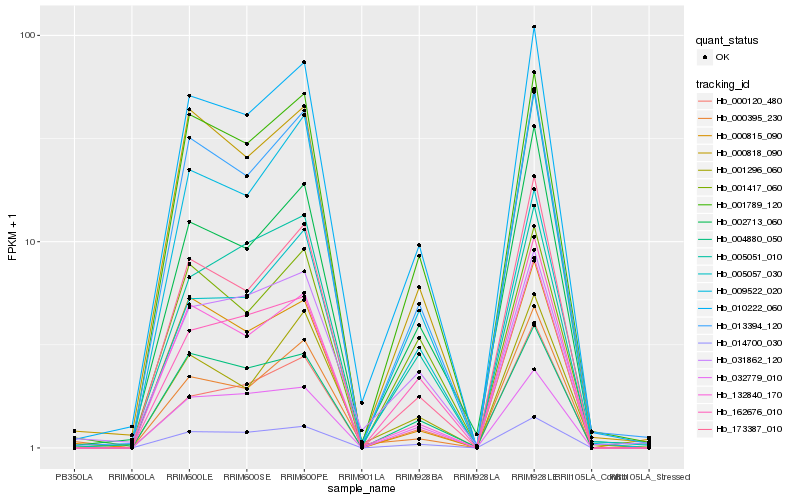
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010222_060 |
0.0 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_013394_120 |
0.0529954366 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 3 |
Hb_014700_030 |
0.0607024585 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 4 |
Hb_001789_120 |
0.0616302494 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_009522_020 |
0.0619680862 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 6 |
Hb_132840_170 |
0.065980458 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 7 |
Hb_002713_060 |
0.0836692678 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 8 |
Hb_173387_010 |
0.0838823505 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 9 |
Hb_004880_050 |
0.085215254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000818_090 |
0.0909541926 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 11 |
Hb_000120_480 |
0.094279154 |
- |
- |
- |
| 12 |
Hb_001296_060 |
0.1009310822 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_005051_010 |
0.1023348854 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_031862_120 |
0.1060225071 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 15 |
Hb_032779_010 |
0.1084254613 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_005057_030 |
0.1088187381 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_001417_060 |
0.1090917947 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 18 |
Hb_162676_010 |
0.1097163647 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000815_090 |
0.1097770936 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000395_230 |
0.1106723407 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Vitis vinifera] |