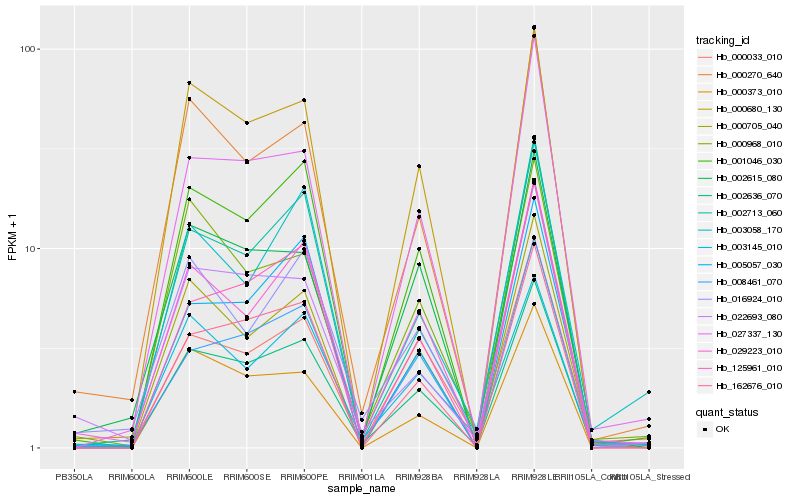
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002636_070 |
0.0 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 2 |
Hb_029223_010 |
0.0812452345 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 3 |
Hb_000680_130 |
0.0874007327 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 4 |
Hb_002713_060 |
0.088578655 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 5 |
Hb_000705_040 |
0.0893520149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000270_640 |
0.0902684821 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 7 |
Hb_027337_130 |
0.0931337159 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 8 |
Hb_000033_010 |
0.0976336891 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 9 |
Hb_008461_070 |
0.0995220305 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_162676_010 |
0.1016240459 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_003145_010 |
0.1053258965 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 12 |
Hb_002615_080 |
0.1076546068 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 13 |
Hb_000373_010 |
0.1124156871 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_016924_010 |
0.1128709799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_022693_080 |
0.1136894851 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 16 |
Hb_125961_010 |
0.1139098036 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_003058_170 |
0.1147449487 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005057_030 |
0.1151504295 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_000968_010 |
0.117608158 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001046_030 |
0.1178306102 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |