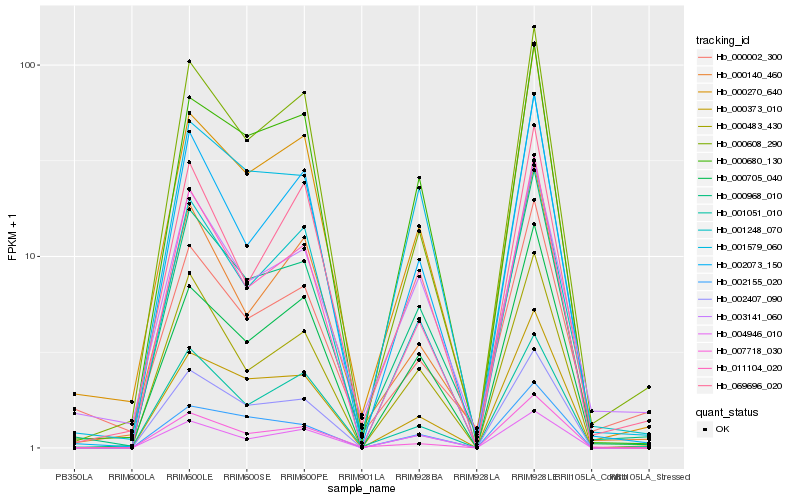
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000968_010 |
0.0 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000270_640 |
0.0842916181 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 3 |
Hb_003141_060 |
0.0853533283 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 4 |
Hb_011104_020 |
0.0900983592 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 5 |
Hb_000680_130 |
0.0944873383 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_001579_060 |
0.0948239993 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 7 |
Hb_000373_010 |
0.0950545629 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_002073_150 |
0.0959014404 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 9 |
Hb_001248_070 |
0.0959285336 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 10 |
Hb_000705_040 |
0.0980686836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000608_290 |
0.1019079114 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 12 |
Hb_000483_430 |
0.1024368801 |
- |
- |
- |
| 13 |
Hb_000140_460 |
0.1060602697 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 14 |
Hb_002155_020 |
0.1078752145 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 15 |
Hb_007718_030 |
0.1104112054 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_001051_010 |
0.1110033204 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 17 |
Hb_000002_300 |
0.1120031576 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_002407_090 |
0.1131134188 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 19 |
Hb_069696_020 |
0.1162818495 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_004946_010 |
0.1164155168 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |