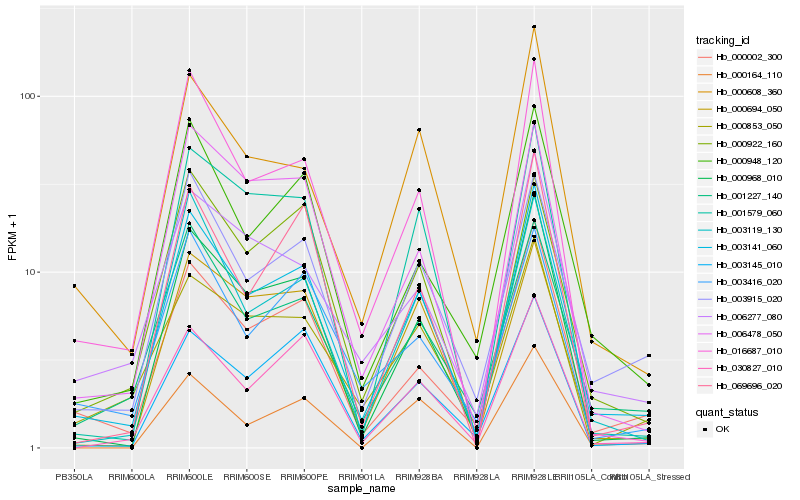
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003141_060 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000968_010 |
0.0853533283 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001227_140 |
0.096404927 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 4 |
Hb_003915_020 |
0.0979665982 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000002_300 |
0.0987981612 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_016687_010 |
0.1051005899 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 7 |
Hb_000948_120 |
0.1060071889 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003119_130 |
0.106288939 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 9 |
Hb_000608_360 |
0.1109557749 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 10 |
Hb_030827_010 |
0.1117951655 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 11 |
Hb_000922_160 |
0.1188492917 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 12 |
Hb_069696_020 |
0.1250544471 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001579_060 |
0.1257347877 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 14 |
Hb_006478_050 |
0.1265551957 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003416_020 |
0.1274709673 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 16 |
Hb_006277_080 |
0.1276765426 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 17 |
Hb_000164_110 |
0.1281558958 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 18 |
Hb_003145_010 |
0.1293440059 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 19 |
Hb_000853_050 |
0.1293991129 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000694_050 |
0.1301138441 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |