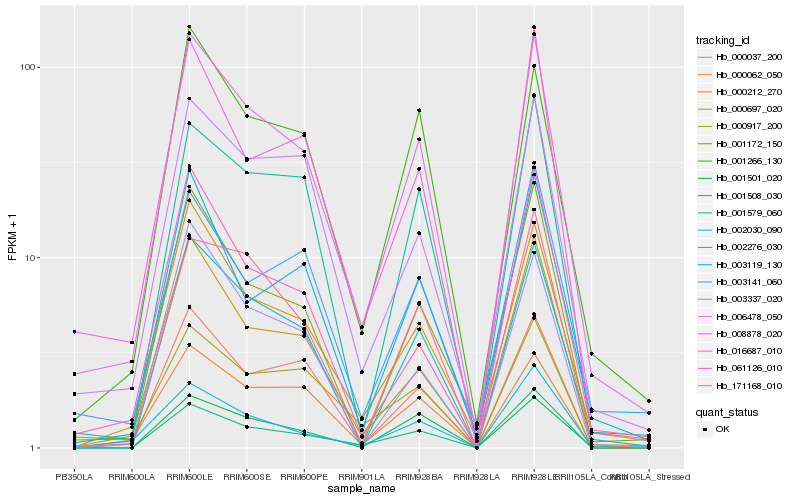
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008878_020 |
0.0 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 2 |
Hb_000212_270 |
0.0809009739 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 3 |
Hb_002030_090 |
0.0920606657 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 4 |
Hb_000697_020 |
0.0993957669 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_001172_150 |
0.0994992077 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 6 |
Hb_001266_130 |
0.1105956265 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 7 |
Hb_006478_050 |
0.1115934406 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_016687_010 |
0.1126279535 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 9 |
Hb_001508_030 |
0.1137225509 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 10 |
Hb_000037_200 |
0.1177221319 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_001579_060 |
0.1184913532 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 12 |
Hb_002276_030 |
0.1192029393 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 13 |
Hb_003119_130 |
0.1203536653 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 14 |
Hb_000062_050 |
0.1213783613 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_171168_010 |
0.1235998018 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_061126_010 |
0.1268822148 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 17 |
Hb_000917_200 |
0.1274444661 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 18 |
Hb_003337_020 |
0.1276711009 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 19 |
Hb_003141_060 |
0.1303321965 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 20 |
Hb_001501_020 |
0.1311976206 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |