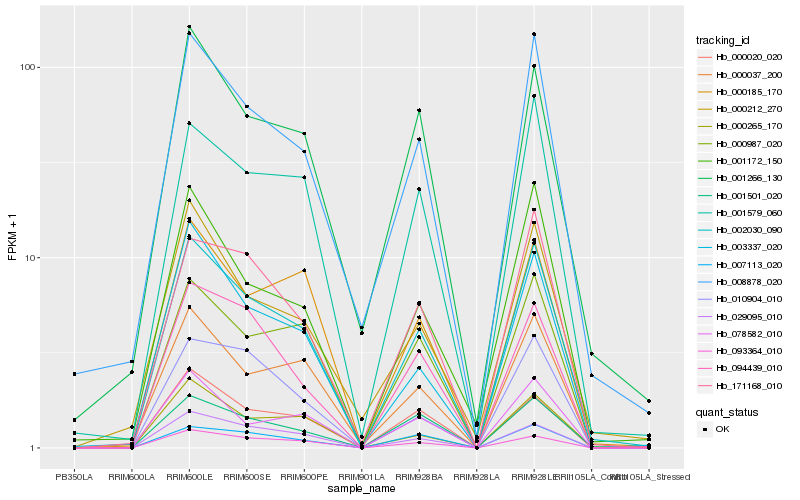
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_090 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 2 |
Hb_008878_020 |
0.0920606657 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 3 |
Hb_093364_010 |
0.1028997364 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 4 |
Hb_001579_060 |
0.1034549329 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 5 |
Hb_000987_020 |
0.1044391072 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 6 |
Hb_000037_200 |
0.1077124311 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_029095_010 |
0.1086410815 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 8 |
Hb_000265_170 |
0.1087815644 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_003337_020 |
0.1103902514 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 10 |
Hb_171168_010 |
0.110958359 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001172_150 |
0.1119681424 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 12 |
Hb_078582_010 |
0.1129415818 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 13 |
Hb_000185_170 |
0.1135782883 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_001501_020 |
0.1140860388 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 15 |
Hb_007113_020 |
0.1179911454 |
- |
- |
- |
| 16 |
Hb_094439_010 |
0.1208256343 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 17 |
Hb_000212_270 |
0.1208947985 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 18 |
Hb_000020_020 |
0.1211060376 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 19 |
Hb_001266_130 |
0.1240700413 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 20 |
Hb_010904_010 |
0.1261971648 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |