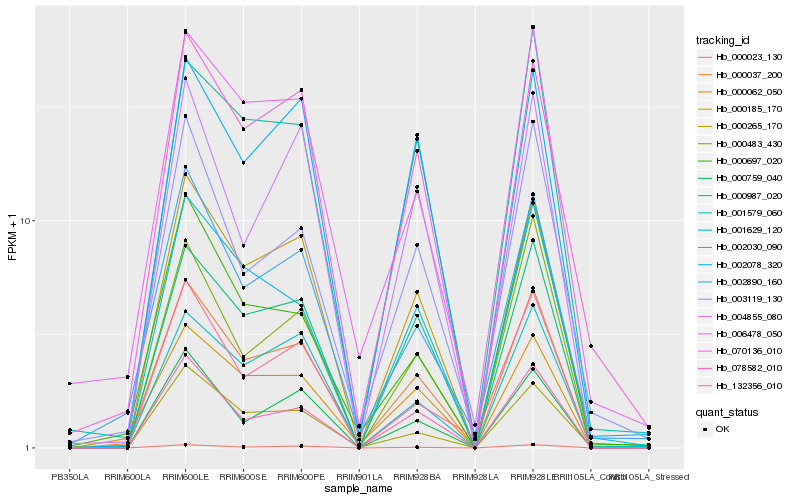
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000037_200 |
0.0 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000185_170 |
0.0693714087 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000062_050 |
0.0768243977 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_078582_010 |
0.0845253362 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 5 |
Hb_000023_130 |
0.0848528325 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 6 |
Hb_000987_020 |
0.088556211 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 7 |
Hb_002890_160 |
0.0960122642 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000265_170 |
0.0963674581 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_006478_050 |
0.1005679153 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_070136_010 |
0.1019611922 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 11 |
Hb_000759_040 |
0.1024521298 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 12 |
Hb_001629_120 |
0.1045201677 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 13 |
Hb_002030_090 |
0.1077124311 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 14 |
Hb_001579_060 |
0.1088573035 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 15 |
Hb_002078_320 |
0.109943013 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_003119_130 |
0.1117920213 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 17 |
Hb_000697_020 |
0.1121474658 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_132356_010 |
0.1136991704 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 19 |
Hb_000483_430 |
0.1156480089 |
- |
- |
- |
| 20 |
Hb_004855_080 |
0.1164352478 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |