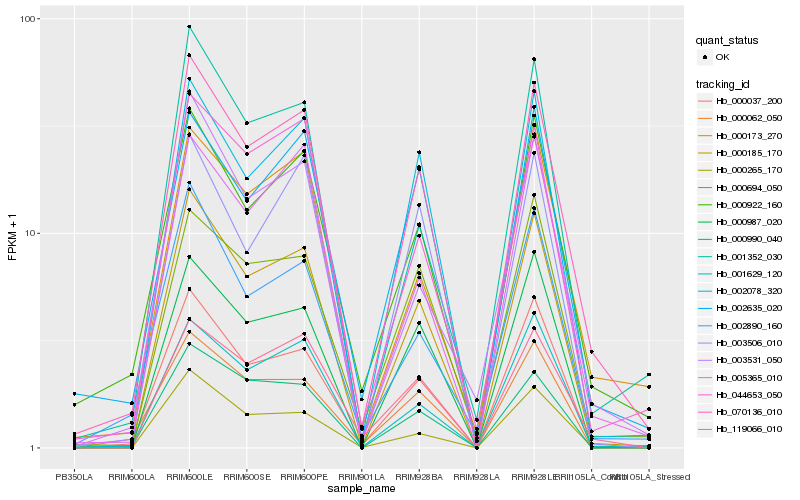
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070136_010 |
0.0 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 2 |
Hb_000185_170 |
0.0637706596 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000922_160 |
0.0896212829 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 4 |
Hb_002078_320 |
0.0906655104 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_119066_010 |
0.0955229102 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 6 |
Hb_000037_200 |
0.1019611922 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001629_120 |
0.1024663378 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 8 |
Hb_002635_020 |
0.10755735 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 9 |
Hb_044653_050 |
0.1078191179 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000173_270 |
0.1109959455 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005365_010 |
0.1110438846 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 12 |
Hb_000987_020 |
0.1114572532 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 13 |
Hb_000990_040 |
0.1121624702 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 14 |
Hb_000062_050 |
0.1127956499 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000265_170 |
0.1136735884 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_000694_050 |
0.1147086962 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 17 |
Hb_003506_010 |
0.1168126326 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 18 |
Hb_002890_160 |
0.1169806078 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003531_050 |
0.1172575536 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001352_030 |
0.1181612872 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |