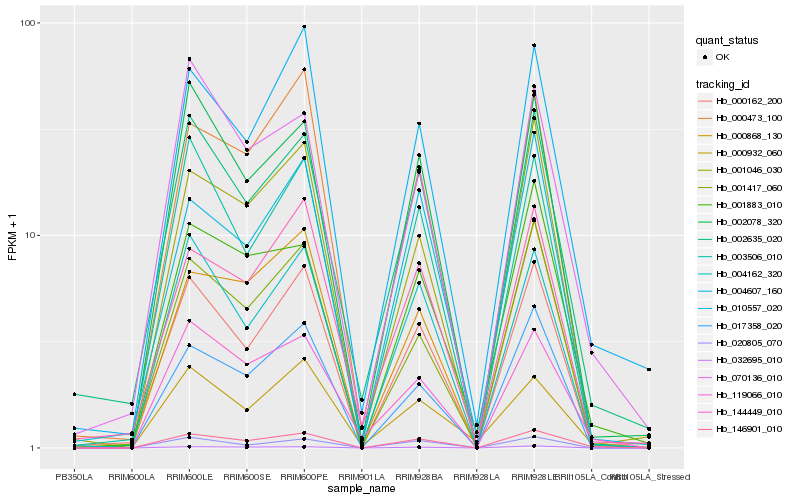
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146901_010 |
0.0 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 2 |
Hb_000162_200 |
0.1068312866 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 3 |
Hb_119066_010 |
0.1128832548 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 4 |
Hb_010557_020 |
0.1153047208 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 5 |
Hb_002635_020 |
0.1184615872 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 6 |
Hb_004162_320 |
0.1191114435 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001883_010 |
0.1198626064 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 8 |
Hb_002078_320 |
0.121703899 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_032695_010 |
0.1217144099 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 10 |
Hb_017358_020 |
0.1246498993 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 11 |
Hb_000868_130 |
0.1261107307 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 12 |
Hb_001417_060 |
0.1274671741 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 13 |
Hb_003506_010 |
0.1290558995 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 14 |
Hb_020805_070 |
0.1294832532 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 15 |
Hb_001046_030 |
0.1303007026 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_004607_160 |
0.1305228067 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 17 |
Hb_000932_060 |
0.1341520264 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_070136_010 |
0.1359567197 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_144449_010 |
0.1370008565 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 20 |
Hb_000473_100 |
0.1383373707 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |