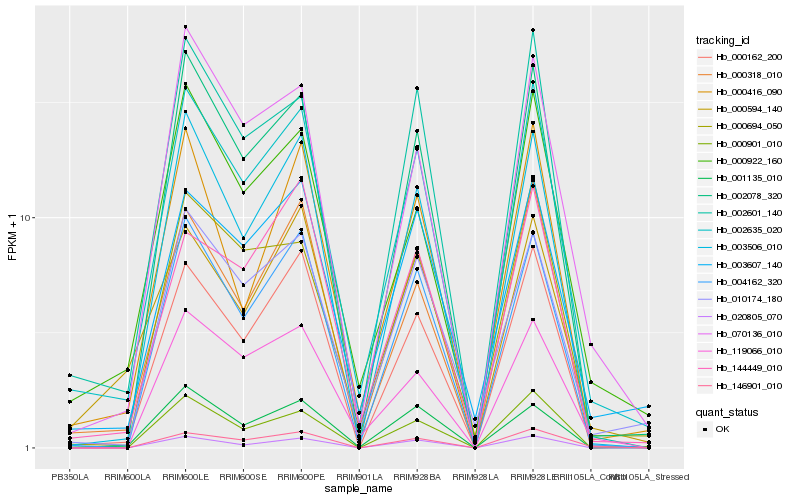
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002635_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 2 |
Hb_002601_140 |
0.0888718226 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 3 |
Hb_000922_160 |
0.0924896283 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 4 |
Hb_002078_320 |
0.093929581 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_000162_200 |
0.0960822444 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004162_320 |
0.097857969 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003506_010 |
0.0996115667 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 8 |
Hb_003607_140 |
0.1008438932 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 9 |
Hb_000318_010 |
0.1013264128 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 10 |
Hb_000694_050 |
0.1056102525 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 11 |
Hb_010174_180 |
0.1058584935 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_070136_010 |
0.10755735 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 13 |
Hb_119066_010 |
0.112200713 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 14 |
Hb_000901_010 |
0.1142635199 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 15 |
Hb_001135_010 |
0.1144412185 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 16 |
Hb_146901_010 |
0.1184615872 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 17 |
Hb_000416_090 |
0.1185735156 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 18 |
Hb_020805_070 |
0.1202097549 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 19 |
Hb_000594_140 |
0.122243039 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 20 |
Hb_144449_010 |
0.1232484584 |
- |
- |
Potassium transporter, putative [Ricinus communis] |